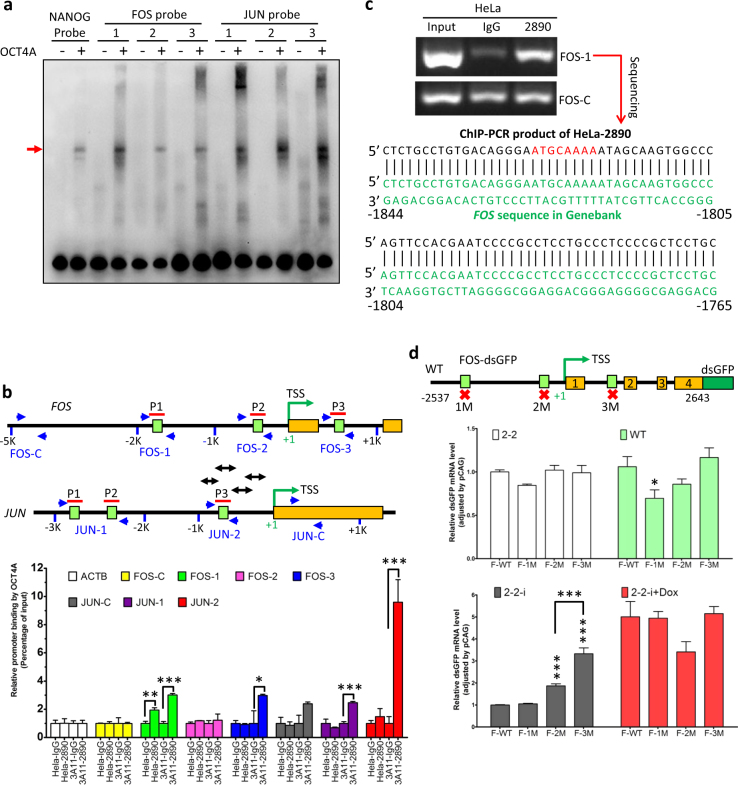
Fig. 5. OCT4A was a transcription factor for FOS/AP-1 in somatic cancer cells.
a EMSA of three sets of biotinylated FOS and JUN probes (relative position shown in (b)), incubated respectively with recombinant His-OCT4A proteins. Biotinylated NANOG probe was applied as a positive control and the red arrow indicated the OCT4A-bound probes. b ChIP-qPCR analysis of WT and 3A11 HeLa cells with anti-OCT4A (CST 2890) and the indicated primers. Upper and middle panel: the schematic representation showing relative positions of predicted octamer motifs (the green boxes), probes (the red lines) and primers (the paired blue arrows) in human FOS (upper) and JUN (middle) genes. Double-headed arrows represent the four DNA fragments of promoter region of JUN covered by primers used by Kuo et al.42. Lower panel: ChIP-qPCR results quantified as relative promoter binding by OCT4A. c Anti-OCT4A (CST 2890)-based ChIP-PCR analysis of HeLa cells with the FOS-1 primers (upper panel). The ChIP-PCR products of HeLa-2890 were sequenced and the partial sequence (black) was aligned with FOS regulatory sequence in public database (green). The putative octamer motif is marked in red. d FOS-dsGFP reporter gene assay of cells with various OCT4A protein expression levels. Upper panel: Schematic representation of the components of normal or single octamer motif mutated FOS-dsGFP reporter plasmids. Middle and lower panels: 2-2, WT, 2-2-i and 2-2-i + Dox were transfected with the wild type or mutated FOS-dsGFP reporter plasmids (F-WT/1M/2M/3M) and the transcription of the reporter genes was evaluated by qRT-PCR. B2M was used as internal control and the relative dsGFP expression was normalized by pCAG primer. The data in (b) and (d) were expressed as mean ± S.D. and mean ± S.E.M., respectively. Two-tailed unpaired Student’s t tests were used. Unless specified, *P < 0.05, **P < 0.01 and ***P < 0.001 were vs. “F-WT”