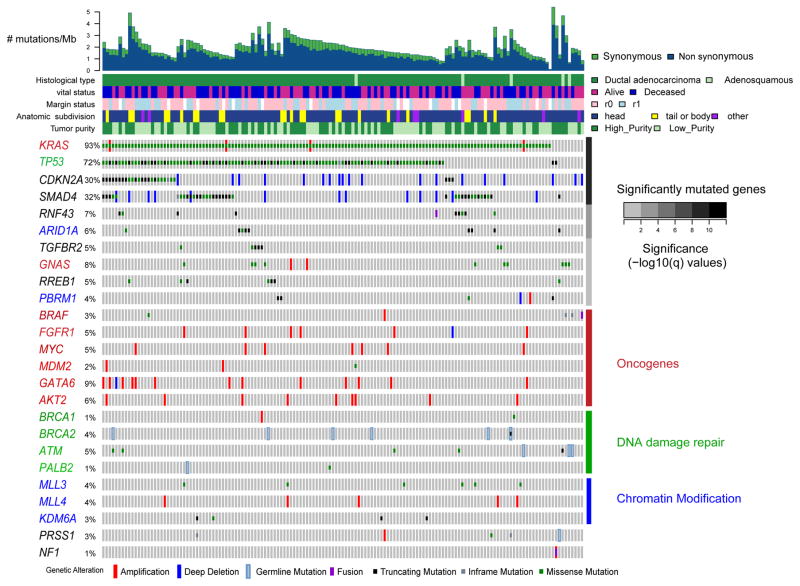
Figure 1. Landscape of genomic alterations in pancreatic ductal adenocarcinoma (PDAC).
Integrated genomic data for 149 non-hypermutated samples (columns), including: mutations (classified as truncating, in-frame or missense); high-level amplifications and homozygous deletions (“Deep Deletion”), fusions derived from analysis of mRNA data, and germline mutations for selected genes as described in the text. Overall number of mutations/Mb and clinicopathologic data for each sample are shown as tracks at the top. Significantly mutated genes (q ≤ 0.1) from exome sequencing data listed in order of q-value, followed by other recurrently altered genes organized in functional classes of oncogenes (red), DNA damage repair genes (green) and chromatin modification genes (blue). Significantly mutated genes from these classes are also colored accordingly. The percentage of PDAC samples with an alteration of any type is noted at the left. See also Figure S1, Tables S1–S3.