Fig. 2. Genome doubling as a function of TP53 aberrations.
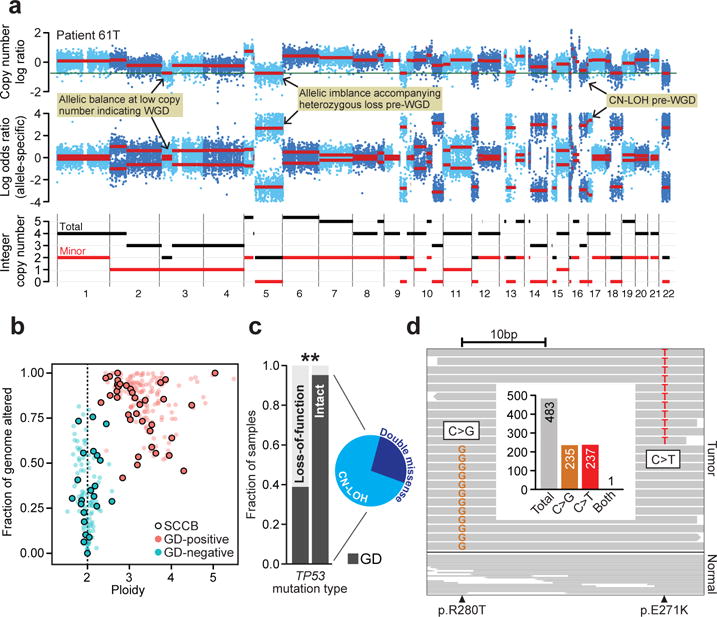
a) Total, allele-specific, and integer copy number segmentation inferred from whole-exome sequencing (chromosomes 1-22; top, middle, bottom) of a representative SCCB genome is shown (patient 61T). Hallmark lesions are indicated (yellow). b) The overall burden of CNAs as a function of tumor ploidy in both SCCBs and urothelial carcinomas (points are individual tumors, SCCBs are those with black border). GD-positive tumors are in red. GD-positive tumors have both higher ploidy and elevated levels of CNAs genome-wide. c) GD as a function of TP53 mutational and zygosity status (biallelic missense with inset representing underlying mechanism; asterisk, p-value < 10−4, Fisher exact test). d) A representative tumor possessing two independent TP53 mutations (patient 70, R280T and E271K) that could be phased, reads spanning both mutant sites indicate that the mutations were in trans. Inset, targeting sequencing coverage of reads spanning both mutations are shown indicating the near absence of reads harboring both mutant alleles.
