Figure 2. Memory-precursor CD8 T cells acquire genome-wide effector-associated DNA methylation programs.
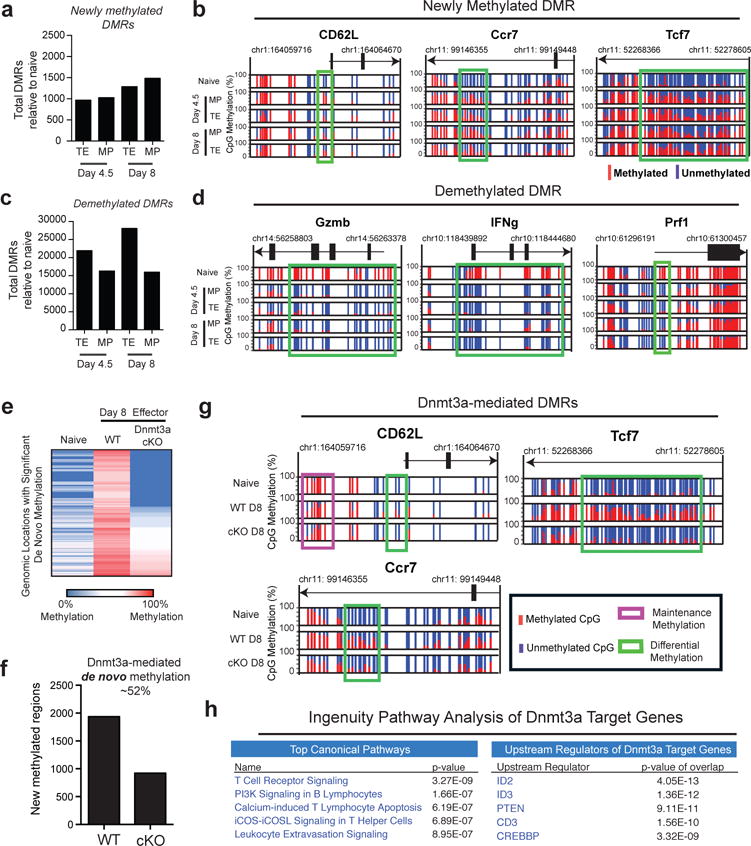
A) Summary graph of the number of newly methylated DMRs in TE and MP subsets relative to naïve cells identified from WGBS analyses. B) Normalized graph of CpG methylation in the Ccr7, Tcf7, and CD62L loci from WGBS data sets. Each vertical line indicates a CpG site and the ratio of red to blue indicate the % of methylated versus unmethylated CpGs at these sites. C) Summary graph of the number of demethylation DMRs between the effector subsets and naïve cells. D) Normalized graph of methylation at CpG sites Gzmb, IFNg, and Prf1 loci from TE and MP WGBS data sets. E) Heat-map representation of top 3000 newly methylated regions (relative to naïve CD8 T cell methylation) from WGBS analysis of tetramer+ WT and Dnmt3a cKO effector CD8 T cell. F) Summary graph of de novo methylated regions in WT and Dnmt3a cKO effector. G) Normalized graph of Dnmt3a-mediated de novo methylation at CpG sites in the CD62L, Ccr7, and Tcf7 loci. H) Top Canonical Pathways and Upstream Regulators from Ingenuity Pathway Analysis of gene-associated Dnmt3a-mediated DMRs.
