Extended Data Figure 5. Conditional deletion of Dnmt3a in activated CD8 T cells inhibits effector-associated de novo DNA methylation but does not impair maintenance methylation.
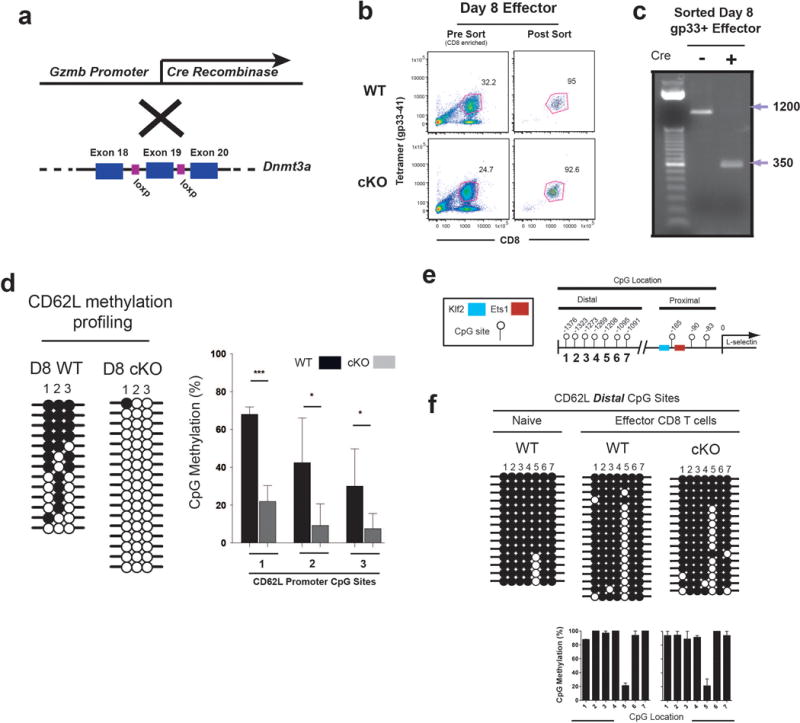
A) Cre recombinase expression is driven by the granzyme b promoter to initiate recombination of Dnmt3a exon 19 following T cell activation. B) Representative FACS analysis of virus-specific CD8 T cells sorted at 8 days post acute viral infection of WT and Dnmt3a cKO mice. C) Recombination of genomic DNA from FACS purified Dnmt3a cKO cells virus-specific CD8 T cells was assessed by PCR using primers that anneal to DNA outside of the floxed target region. The larger PCR amplicon corresponds to the intact locus and the smaller PCR product is the amplicon of the recombined locus. D) Representative and graphical summary of CD62L promoter methylation in WT and Dnmt3a cKO cells. Average and standard deviation were calculated from bisulfite sequencing analysis of 6 individually sorted populations. E) Cartoon diagram of CD62L promoter CpG location proximal and distal to the transcriptional start site. F) Representative DNA methylation analysis of CpG sites distal to the CD62L promoter regions in Day 8 WT and Dnmt3a cKO antigen-specific effector CD8 T cells. Graphical summary of the average CD62L distal CpG methylation in WT and Dnmt3a cKO cells calculated from bisulfite sequencing analysis of 4 individually sorted populations.
