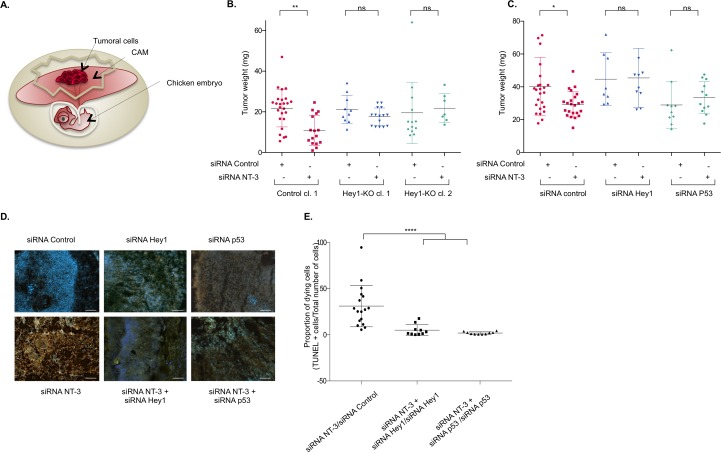
Fig 6. p53 and Hey1 are essential for TrkC-mediated inhibition of tumoral growth in a xenograft model of neuroblastoma.
(A) Schematic representation of the experimental setting of tumor growth in the chick embryo. Tumoral cells are inoculated on the CAM of E10 chick embryo. Five d later, a primary tumor has grown and can be dissected, weighted, and immunohistologically analyzed. (B) Measures of tumor weights 5 d after inoculation of Control (cl.1) and Hey1-KO (cl.1 and cl.2) SHEP clones generated by CRISPR/CAS9 editing and transfected with siRNA control or siRNA NT-3. A one-way ANOVA with Sidak’s multiple comparison test is performed (siRNA NT-3 versus siRNA). **p < 0.01. (C) Measures of tumor weights for each condition, as indicated. A one-way ANOVA with Sidak's multiple comparison test is performed (siRNA NT-3 versus siRNA control, siRNA NT-3 + siRNA Hey1 versus siRNA Hey1, siRNA NT-3 + siRNA p53 versus siRNA p53), taking into account siRNA intrinsic toxicity of siRNA p53. *p < 0.05. (D) TUNEL (orange) and DAPI (blue) staining performed on cryosections of the dissected tumors from (C). A representative picture is shown for each condition. ×20 magnification, scale bar = 1 μm. (E) TUNEL-positive cells and DAPI-positive cells were quantified by ImageJ64 for n = 3–8 sections of at least 3 tumors for each condition. The ratio of TUNEL-positive cells over the total number of cells (DAPI positive) is calculated. A ratio with the value obtained in the corresponding control condition (siRNA NT-3 versus siRNA control, siRNA NT-3 + siRNA Hey1 versus siRNA Hey1, siRNA NT-3 + siRNA p53 versus siRNA p53) is calculated as indicated for each section to consider siRNA intrinsic toxicity, and the mean values ± SEM (n = 3) are indicated. A 2-sided Mann-Whitney t test is performed, ****p < 0.0001. Underlying data can be found in S1 Data. CAM, chorioallantoic membrane; CAS9, CRISPR-associated protein 9; CRISPR, clustered regularly interspaced short palindromic repeat; KO, knock-out; ns, nonsignificant; NT-3, neurotrophin-3; siRNA, small interfering RNA; TrkC, tropomyosin receptor kinase C.