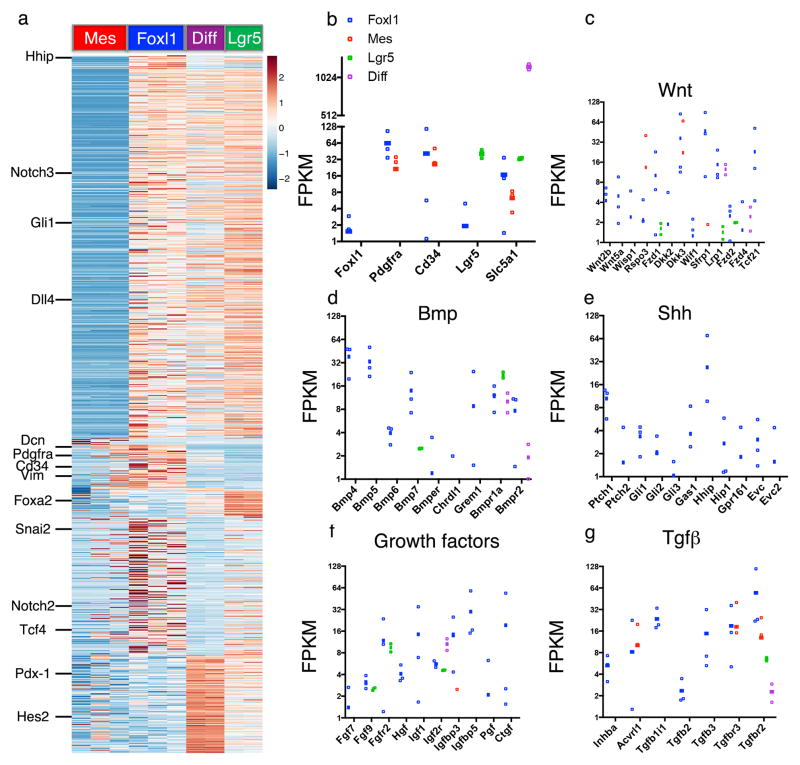
Figure 2. Telocytes express key signaling molecules.
RNAseq analysis of Foxl1+ cells (n=3 biologically independent animals) compared to Foxl1− mesenchymal cells (n=3 biologically independent animals), Lgr5+ stem cells (n=2 biologically independent animals), and differentiated enterocytes (n=2 biologically independent animals). (a) Heatmap showing hierarchical clustering. Only genes with a fold-change > 10 in pairwise comparisons and an FDR < 2% were used for the analysis. Values are normalized z-scores, plus or minus standard deviation. (b–g) Selected markers showing differential expression: (b) Foxl1, Pdgfra, Cd34 (telocyte markers), Lgr5 and Slc5a1, (c) Wnt pathway, (d) Bmp pathway, (e) Shh pathway, (f) Growth factors, (g) Tgfβ pathway. Data are shown as FPKM. Error bars indicate SEM.