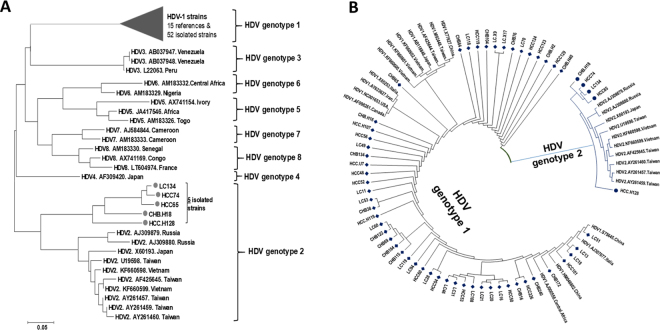
Figure 2.
Phylogenetic analysis of isolated HDV genotypes. (A) A phylogenetic tree was constructed based on the alignment of 235 bp of 57 nucleotide sequences isolated from HDV/HBV co-infected patients. 39 full-length HDV genomes through HDV1-8 retrieved from NCBI database along with GenBank accession numbers were included for the analysis. A neighbor-joining tree was constructed with a bootstrap of 1000 replicates. The bar at the base of the tree indicates the scale for nucleotide substitutions per position. (B) The phylogenetic tree was constructed only for HDV genotype 1 and 2 sequences and involves 82 nucleotide sequences (25 references of full-length HDV genome retrieved from NCBI database, 52 strains of HDV genotype 1 (denoted as ♦) and 5 HDV genotype 2 (denoted as •) from HBV/HDV coinfected patients in our study group.