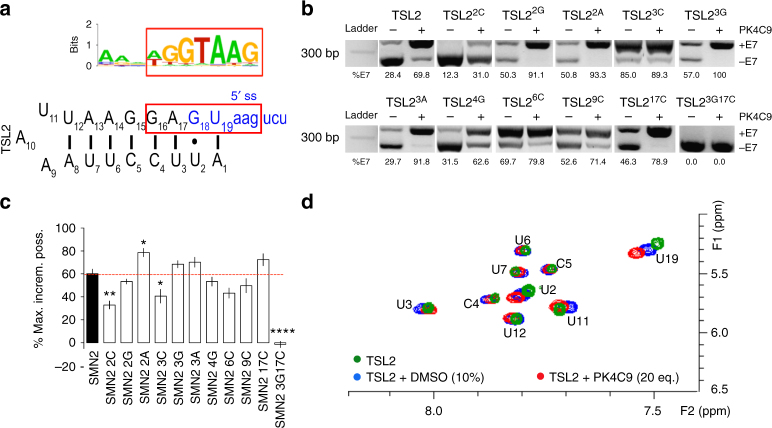
Fig. 5.
PK4C9 binding to TSL2 affects the 5′ ss of E7. a Motif search from RNA-sequencing data (n = 4 biological replicates), using algorithms Gibbs, MEME and Weeder, identified AGGTAAG as the most enriched motif in exons that are differentially spliced-in SMA cells upon treatment with PK4C9 (40 μM, 24 h) compared to SMA cells treated with DMSO (24 h). The illustration shows the example found by Weeder and the similarity with TSL2 (red box). b RT-PCR from HeLa cells transfected with SMN2E6−to−8 minigenes16 carrying n.m. TSL2 or the indicated structural mutations. Transfected HeLa cells were treated with DMSO (0.04%, controls) or PK4C9 (40 μM) (n = 9; three biological and three technical replicates). c The effect of these mutations on PK4C9 activity was measured as the percentage of maximum E7 increment possible (see Eq. 2). d TOCSY NMR spectrum showing the cross-peak region of H5/H6 protons of the pyrimidine bases of TSL2 (40–50 μM) with and without PK4C9 (20 equivalents). 1H chemical shifts were referenced to the internal DMSO signal (2.63 ppm). Residues U2 and U19 showed the most significant chemical-shift perturbances. *p < 0.05, **p < 0.01 and ****p < 0.0001. p-values were obtained by applying non-paired, two-tailed t tests with Welch corrections. The graph shows mean values ± SEM