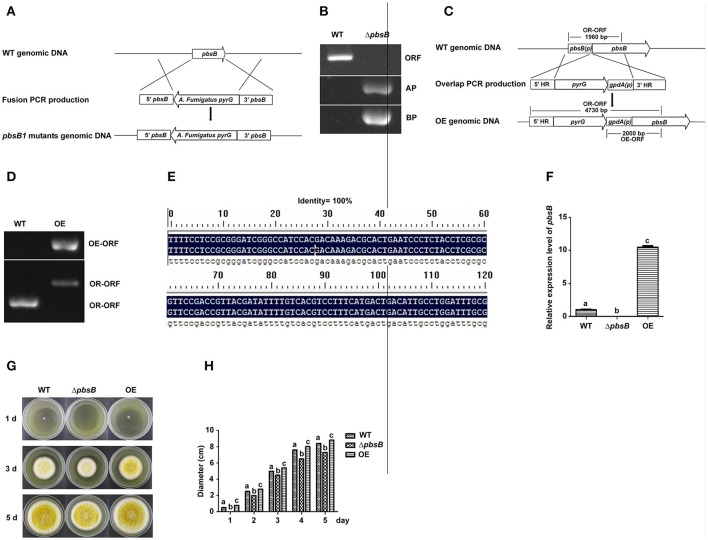
Figure 2.
PbsB involved in the growth of A. flavus. (A) The scheme for ΔpbsB strain construction by homologous recombination. 5′-UTR (5′pbsB) and 3′-UTR (3′pbsB) of pbsB and 1.89 kb pyrG from A. fumigatus were amplified, respectively, and they were fused together with nesting primers. The pbsB deletion strain was prepared with pyrG to replace pbsB in WT (CA14Δku70ΔpyrG) through transformation the protoplast with the fusion production of 5′-UTR-pyrG-3′-UTR by homologous recombination. (B) The results of PCR analysis, in which ORF was amplified with primer pbsB-OrfF and pbsB-OrfR, AP was with pbsB-AF and pyrG-R, and BP was with pyrG-F and pbsB-AR. (C) The scheme for OE strain construction by homologous recombination. (D) OE strain was detected with PCR analysis. OE-ORF was amplified with primer gpdA-F and OE-R, and OR-ORF was with primer OE-overlap-F and OE-R. (E). The alignment of flanking regions of pyrG and 5′ and 3′UTR of pbsB in ΔpbsB strain between sequencing results and constructed sequence map. (F) The expression level of pbsB in WT, ΔpbsB, and OE strain were analyzed with qRT-PCR. (G) Colony of WT A. flavus, ΔpbsB mutant and OE strains were cultured in YES medium for 1 d to 5 d. (H). Comparative analysis of colony diameters, the letters “a,” “b,” and “c” used in the histogram of this study represented the significant difference among WT, ΔpbsB mutant, and OE strain (P < 0.05).