Abstract
Supergenes consist of co-adapted loci that segregate together and are associated with adaptive traits. In the fire ant Solenopsis invicta, two ‘social’ supergene variants regulate differences in colony queen number and other traits. Suppressed recombination in this system is maintained, in part, by a greater than 9 Mb inversion, but the supergene is larger. Has the supergene in S. invicta undergone multiple large inversions? The initial gene content of the inverted allele of a supergene would be the same as that of the wild-type allele. So, how did the inversion increase in frequency? To address these questions, we cloned one extreme breakpoint in the fire ant supergene. In doing so, we found a second large (greater than 800 Kb) rearrangement. Furthermore, we determined the temporal order of the two big inversions based on the translocation pattern of a third small fragment. Because the S. invicta supergene lacks evolutionary strata, our finding of multiple inversions may support an introgression model of the supergene. Finally, we showed that one of the inversions swapped the promoter of a breakpoint-adjacent gene, which might have conferred a selective advantage relative to the non-inverted allele. Our findings provide a rare example of gene alterations arising directly from an inversion event.
Keywords: supergene, inversion, social chromosome, fire ant, Solenopsis invicta, polygyne
1. Introduction
Supergenes often consist of multiple co-adapted loci that segregate as a single unit, and importantly, are associated with polymorphism for complex adaptive traits [1–4]. Classic examples of supergenes include butterfly mimicry [5]; plant self-incompatibility and heterostyly [6,7]; and meiotic drive systems [8–10], such as the t-locus in mice [11] and segregation distorter in Drosophila [12]. With the aid of genomic technologies, supergenes have recently been demonstrated in additional systems, including ants [13,14], birds [15–17], butterflies [18,19], fishes [20] and plants [21].
In the fire ant, Solenopsis invicta, a supergene regulates queen odour, worker behaviour and a suite of other traits resulting in colonies having only one queen or many queens [13,22–24]. This ‘social’ supergene was identified by genetic mapping as a region lacking recombination in heterozygous queens bearing the two alternate alleles, SB and Sb, named after the associated Gp-9 alleles [13,25,26]. Recombination within the supergene occurs normally in SB/SB queens and could occur in Sb/Sb queens but does not because of the absence of functional Sb/Sb queens at least in the invasive range [24,26]. The supergene was estimated to be at least 12.5 Mb long with approximately 600 genes, and is probably larger based on a subsequent molecular evolution analysis [27]. The lack of recombination is probably owing in part to a large inversion (greater than 9 Mb) [13].
The goal of this study was to understand the evolution of the supergene in the fire ant S. invicta. Although the locations of the fire ant social supergene boundaries are approximately known from genetic map and molecular evolution analysis [27], the exact inversion breakpoints are still unknown. Finding the breakpoints is useful for delimiting the supergene boundary. Additionally, a comparison of the breakpoints between the SB and Sb alleles can reveal if there is one simple, major inversion or several large ones. Finally, a breakpoint may occur within or near a gene, creating a mutation or rewiring gene regulation; such a gene would be a candidate gene affecting fire ant social form differences. If beneficial, such a mutation could help explain how the frequency of a new inversion allele increased in a population.
In this study, we report the cloning of multiple boundary breakpoints in the fire ant supergene. The first was found fortuitously by a bacterial artificial chromosome (BAC) spanning the breakpoint. Subsequent comparisons of the genetic map and genomic assemblies revealed one major inversion and at least one additional large (greater than 800 Kb) rearrangement. Furthermore, we determined the temporal order of the two big inversion events based on the translocation pattern of a small fragment. Finally, we found that the inversion has caused the swap of a putative enhancer or promoter, changing the gene expression levels of a gene located at one breakpoint. The results advance our understanding of the evolutionary history of the fire ant supergene and provide a rare example of gene alterations arising directly from an inversion event.
2. Material and methods
(a). Ant samples and genotyping
Colonies were collected from Taoyuan, Taiwan. The red imported fire ant, S. invicta, from Taiwan used in this study is not endangered or protected. Ant husbandry followed Academia Sinica regulations.
A preliminary assignment of monogyne or polygyne social form was based on mound structure and distribution in the field as well as queen number and worker size distributions in the laboratory. Subsequently colony social form was validated by genotyping at the Gp-9 locus using a polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP) assay [25] on the genomic DNA from a mixture of 10 adult workers from a colony. We obtained male larvae from orphan colonies where dealated queens were producing haploid males. For validation of the breakpoint joins by PCR, adult male DNA from monogyne or polygyne colonies was used. Males were genotyped at the Gp-9 locus as well [25].
(b). Bacterial artificial chromosome-fluorescence in situ hybridization
We had previously end-sequenced random clones from the SW_Ba BAC library (Clemson University Genomics Institute, South Carolina, USA) and identified several BACs that mapped to the social chromosome (electronic supplementary material, table S1) [28]. Clone A18 from plate 073 was used in a previous study [13] and is located within the non-recombining supergene region. Clone M24 from plate 145, identified in this study, covers the ‘right’ breakpoint of the social chromosome.
BAC-fluorescence in situ hybridization (FISH) experiments were conducted as previously described [13,29]. In brief, we grew an overnight 20 ml culture of each clone and then purified BAC DNA using the Plasmid Mini Kit (12125, QIAGEN). BAC DNA was then labelled with fluorescent dyes (A18: Alexa Fluor 488; M24: Alexa Fluor 647) by nick translation with the FISH Tag™ DNA Multicolor Kit (F32951, ThermoFisher). Chromosome spreads were prepared using a standard ant protocol [30] from testes of fourth instar male larvae. Then, slides were hybridized with the labelled BAC probe overnight (greater than 12 h) at 37°C. After hybridization, slides were washed three times with 62°C pre-warmed washing buffer (0.1× SSC, 1% TritonX-100), and then rinsed briefly with 2× saline-sodium citrate (SSC) and double distilled water (ddH2O) to remove residual salt and detergent. Chromosomes were mounted in the VECTASHIELD mounting medium with 4′6-diamidino-2-phenylindole (DAPI) (H-1200, Vector Laboratory). Images were captured with the DeltaVision Microscopy System and processed by deconvolution to improve image resolution. A18 (green) and M24 (red) signals were false-coloured and merged with DAPI images using Photoshop software.
Primers were designed using Primer3 [31] and their sequences are shown in the electronic supplementary material, table S2. Seven breakpoint joins (two SB-specific: J1 and J2; five Sb-specific: J3 to J7) were detected by conducting PCR analysis on each of three independent SB and Sb male samples. The PCR reactions were performed with Taq PCR MasterMix (KTT-BB01M, Tools) in 10 µl volumes containing 5 ng of total DNA and 0.2 µM forward and reverse primers. PCR amplifications were performed with the following profile: initial 4 min denaturation at 94°C; followed by 35 cycles of 30 s denaturation at 94°C, 30 s annealing at 60°C and 1 m 10 s elongation at 72°C; and a final 10 min extension at 72°C.
The diagnostic approximately 600 bp fragment (C) along with approximately 500 bp upstream and downstream flanking sequences from the SB and Sb genomes were amplified with the HiFi PCR Kit (KK2103, KAPA), and then cloned with the Zero Blunt TOPO PCR Cloning Kit (450245, ThermoFisher) for Sanger sequencing.
(c). Genome comparisons
The six Sb contigs used in this study are available under NCBI accession MH121689 to MH121694 and the Dryad repository (doi:10.5061/dryad.2458p4r) [28]. They were derived from an Sb genome assembled using the Falcon assembler from approximately 40× sequence coverage using the PacBio platform [32]; the full genome will be reported later. We used blastn and Artemis Comparison Tool (ACT) [33] for the comparison of the scaffolds between the SB (genome G [34]) and Sb genomes to find potential breakpoints. Sequence matches were filtered (bit score greater than 1400, identity greater than 90%) and displayed in ACT. To confirm the critical breakpoint joins, we mapped raw PacBio reads to the new Sb assembly using Blasr [35] and then inspected the focal contigs for contiguous reads crossing the join in integrative genomics viewer [36]. We also examined the original Sb contigs for suspicious merges similarly. We found that approximately 53 Kb on the left end of contig 000102F was wrongly merged at a DNA transposon, and thus we trimmed it to yield 000102F-a. Similarly, contig 000181F was incorrectly assembled at a repetitive region, so we split it into two contigs and trimmed the redundant sequences to yield 000181F-1 and 000181F-2.
(d). RNA analysis
The gene expression values for the three genes adjacent to the breakpoints are from a separate antennae gene expression experiment (electronic supplementary material, table S3); this was the only RNA-seq dataset available with separate samples for SB/SB and SB/Sb individuals. In brief, we conducted RNA-seq on four biological replicates of RNA extracted from the worker antennae of three classes: monogyne SB/SB, polygyne SB/SB and polygyne SB/Sb. Reads were mapped to the fire ant genome using BWA-mem with default parameters [37]. Gene expression level was estimated with HTSeq-count [38]. We then used the edgeR software package [39] to look for gene expression differences. Significance values are after multiple test correction using Bonferroni correction (three target genes; nine comparisons) and the Benjamini–Hochberg method (full dataset). To analyse allele-specific expression (ASE), single nucleotide polymorphism (SNPs) and indels in the untranslated region (UTR) and coding regions were called using GATK [40–42] for the worker antennae (pool of four bioreplicates) dataset based on the BWA-mem alignment file. Sb-specific SNPs/indels were identified based on a comparison of low coverage sequence from seven pairs of SB and Sb brothers [13].
We used the official gene set annotation [34] in the main text; their correspondence to the NCBI Gene ID's are: SINV22157 is 105193832; SINV22107 is 105193833; and the fused gene SINV23002 plus SINV23011 is 105199310 in NCBI annotation release 100 [43]. Exon–exon fusion between the last and the first exons of SINV23002 and SINV23011 was confirmed by inspecting RNA-seq reads.
3. Results
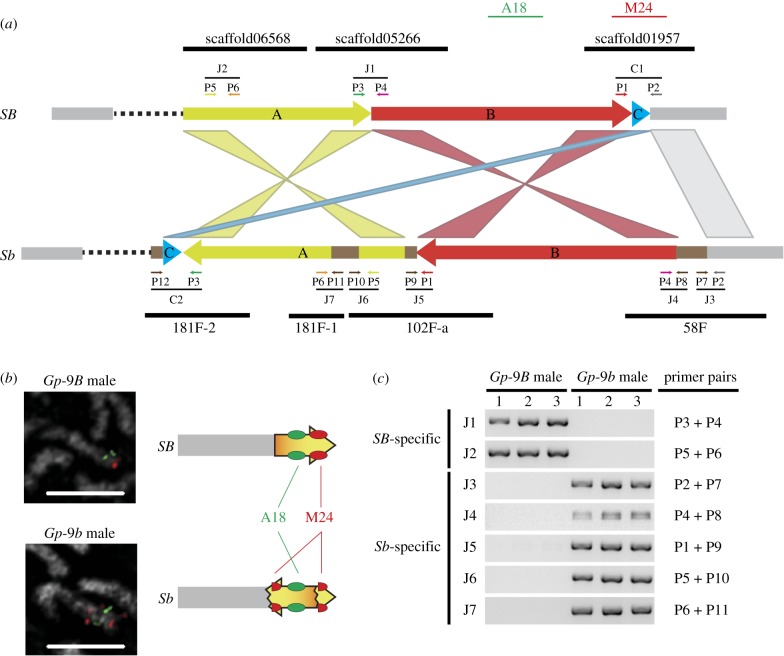
We previously used BAC-FISH to demonstrate the presence of one large inversion in the fire ant supergene [13]. Subsequent examination of addition BACs revealed one (BAC_145M24) with an interesting hybridization pattern. On the SB chromosome there was only one robust hybridization signal (figure 1b and electronic supplementary material, figure S1). In comparison, on the Sb chromosome, we observed two signals that were separated by approximately one-third of the chromosome and flanking another BAC probe known to be within the inversion (figure 1b and electronic supplementary material, figure S1). Since the BAC library was made from SB/SB individuals, this hybridization result suggested that BAC_145M24 might span the right breakpoint on SB.
Figure 1.
The rearrangements on the fire ant social chromosomes. (a) Schematic diagram of the SB and Sb genomic structure around the supergene. Fragments A (yellow), B (red), and C (blue) comprise around three-quarters of the supergene in the SB chromosome. The rest of the supergene (dashed line) and the outside edges (grey boxes) are shown. Sb-specific sequences (brown boxes) in between or within fragments are shown. The simplified alignments between the SB and Sb fragments are shown with corresponding simple (same direction) or twisted (inversion) colour ribbons (see also the electronic supplementary material, figure S2). Informative scaffolds in the SB and Sb genomes are indicated as black lines. The positions of primers (P1 ∼ P12), PCR junctions (J1 ∼ J7) and cloned regions (C1 and C2) shown in c and S3 are indicated. Locations of BAC probes (A18 and M24) in SB are illustrated. (b) BAC-FISH identifies a breakpoint between the SB and Sb social chromosomes. The right panel shows a schematic interpretation of the hybridization patterns owing to the inversion of fragment B. Scale bars, 5 µm. (c) PCR amplifications of the SB- (J1 and J2) or Sb-specific junctions (J3 to J7) with three independent haploid Gp-9B and Gp-9b male samples.
Based on the SB genome (version G), BAC_145M24 is located on BigB_G_scaffold01957 and spans positions 401 094 to 493 803. This is near the ‘right’ (or ‘bottom’) end of the social chromosome on the genetic map [13]. To corroborate that the breakpoint is near this region, we inspected RADseq data based on genetic maps from four polygyne families [13]. We looked for the ‘first’ RADtag exhibiting recombination between the SB and Sb alleles. The most informative RADtag was from family P033 and was located at position 530 490. The breakpoint, therefore, must be before this position which is consistent with the position of BAC_145M24.
To determine the exact breakpoint location, we conducted a BLASTN search [44] to query the approximately 93 kb SB sequence bounded by the two end sequences of BAC_145M24 against a preliminary PacBio assembly of the Sb genome [28]. The corresponding Sb sequence lies on two scaffolds (littleb_000058F and littleb_000102F-a; figure 1a and electronic supplementary material, figure S2) and manual inspection of the alignments localized the breakpoint to position 439 985 on BigB_G_scaffold01957. Additional examination within this BAC region revealed many small indels (insertion/deletions) and one moderate approximately 25 Kb insertion on the Sb scaffold, which appears to be the remnant of one or more transposons [28].
If the SB and Sb alleles of the supergene differed primarily by one large simple inversion, there should be a two-to-two correspondence of scaffolds associated with the respective SB and Sb breakpoints. However, comparing the two Sb scaffolds back to the SB genome revealed three hits: the known right scaffold, BigB_G_scaffold01957; and two ‘left’ scaffolds, BigB_G_scaffold06568 and BigB_G_scaffold05266 (figure 1a and electronic supplementary material, figure S2). The two left scaffolds are near each other on the genetic map (within approximately 2 cM) and BigB_G_scaffold06568 is within approximately 1 Mbp of the left boundary determined from the genetic mapping data.
The three hits suggested a more complicated inversion scenario, so we next conducted a more extensive comparison of the SB and Sb scaffolds. Using the two new SB scaffolds to query against the Sb genome we found that they corresponded to one additional Sb contig (littleb_000181F-2). Furthermore, the corresponding Sb sequence orientation requires one additional inversion of fragment ‘A’ (approx. 844 Kb) (figure 1a). An initial global overview of the inversions seemed to indicate that the ‘main’ inversion (hereafter inversion ‘1’) of fragment ‘B’ (greater than 9 Mb) could have happened independently of this second inversion (2). In other words, the temporal order of the two inversions seemingly appeared unknown.
However, a more detailed inspection revealed an informative approximately 600 bp fragment (C) that uncovers the probable temporal order of the two principle inversions (figure 1). On the SB genome, fragment C is located at the right breakpoint next to fragment B. However, fragment C is no longer directly adjacent to its neighbouring SB sequence in the Sb assembly and is, in fact, translocated to the other end (arrowhead) of fragment A. This translocation was confirmed using PCR and sequencing assays (electronic supplementary material, figure S3). We also validated the other seven additional hypothesized breakpoint joins suggested by the SB and Sb genome sequences (figure 1c). These results suggested that the simplest model for the evolutionary history of the Sb social chromosome is that inversion ‘1’ occurred before ‘2’ (figure 2d).
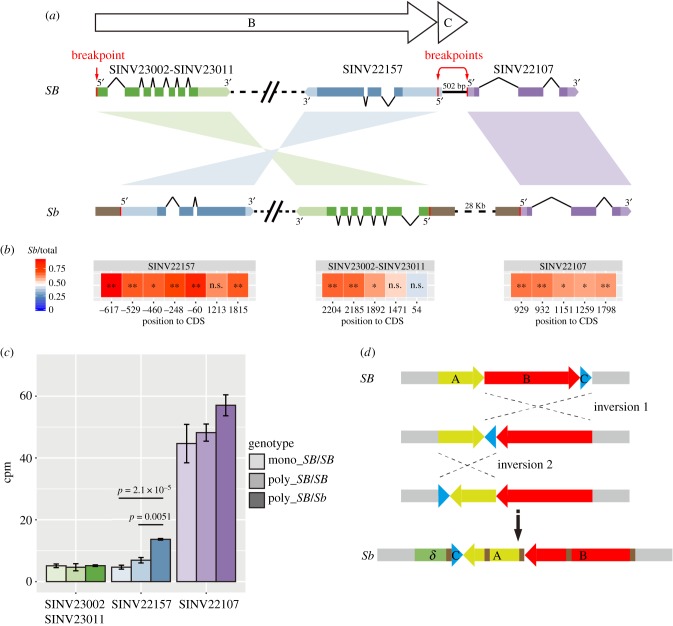
Figure 2.
(a) Breakpoints are present at the 5′-untranslated region (UTR) of three protein-coding genes in Sb. Genes are depicted as boxes with light (UTR) or dark (exon) colours with linked lines. The direction of each gene is shown with 5′ and 3′ labels, accompanied by simple (same direction) or twisted (inverted) ribbons in between the SB and Sb gene pairs. Breakpoints (arrows) and their corresponding joins (red lines) with accumulation of transposons and other sequences (brown rectangles) in Sb are shown. The open arrows illustrate the directions of fragments B and C, as depicted in figure 1a. (b) Allele-specific expression of the three genes in SB/Sb worker antennae. Heat map shows the ratios of Sb-allele-specific reads over total reads for each SNP or indel within the UTR and coding region. Variants are oriented based on the gene direction in Sb, and their nucleotide positions to the coding DNA sequence are shown below. * or ** indicates unadjusted p < 0.05 or <0.01, binomial test; full data in the electronic supplementary material, table S5. (c) Expression levels of the three genes located at the breakpoints in antennae of workers of different genotypes. Gene expression levels are the counts per million mapped reads (cmp) averaged over four bioreplicates for each sample type. P-value scores were determined using edgeR and displayed if they passed a Bonferroni correction α = 0.0056 (nine comparisons). Error bars are standard deviations. Mono, monogyne; poly, polygyne. (d) Proposed scenario for the evolutionary history of the Sb social chromosome. (1) Joint inversion of fragments B and C from the ancestral SB chromosome (inversion 1), contributing to the largest genome rearrangement (greater than 9 Mb) between the SB and Sb social chromosomes. (2) Capture of fragment C (approx. 600 bp) by inversion 2, resulting in an additional 844 Kb rearrangement. (3) Accumulation of repeats, e.g. tandem repeats or transposons and other sequences (brown rectangles) throughout the supergene region of the Sb chromosome owing to the lack of recombination. The conceptual fragment δ is predicted to form a third inversion, together with the rearranged fragments A, B and C, comprise the supergene of the Sb social chromosome. The position and numbers of repeats are illustrative and are only depicted on the final line but may have appeared earlier.
Numerous examples in humans, Drosophila, and other species have indicated that some inversions could be caused by ectopic homologous recombination at transposons or other duplicated sequences [45–50]. We examined if fragment C might be a multicopy fragment but found no evidence for it in either the SB or Sb assemblies. Additionally, in the Solenopsis geminata genome the orthologous fragment C is a single copy sequence [28]. Thus, it seems unlikely that fragment C contributed to the formation of the inversions. We also looked for the presence of transposons in the sequences directly adjacent to each breakpoint. We found that the genomic breakpoints in Sb are often flanked by transposon sequences, but such transposons were not obvious in the SB haplotype. These results suggest that transposons were unlikely to have mediated the inversion events, but perhaps accumulated after the cessation of recombination. Precise estimation of the ages of the inversions and associated transposons will be needed to examine this possibility.
There are four breakpoints associated with the two derived inversions in Sb; therefore, we examined the breakpoints to see if any genes were mutated. Three predicted protein-coding genes (SINV22157, SINV22107 and fused SINV23002–SINV23011) are within 2 Kb of the four breakpoints in the SB assembly (figure 2a). The protein-coding capacities of these genes are intact as none have the breakpoint within the coding region. However, interestingly SINV22157, which encodes an unknown domain-containing protein (DUF4506, pfam14958), was more highly expressed in the worker antennae of SB/Sb versus SB/SB individuals in an RNA-seq dataset (approx. 1.8-fold higher than polygyne SB/SB, p-value = 0.0051; 2.8-fold higher than monogyne SB/SB, p-value=2.1 × 10−5; edgeR F-test on four biological replicates; Bonferroni threshold for nine tests, α = 0.0056; figure 2c; electronic supplementary material, tables S3 and S4). We did not detect differential expression for SINV22107 and SINV23002–SINV23011 in this dataset, although they may have differential expression elsewhere.
Examination of the RNA-seq reads revealed that the breakpoints occur in the 5′-UTR for all three genes. Thus, the transcriptional start sites, including the promoters, are different between the SB and Sb alleles. Fragment C encompasses the entire intergenic region between the 5′ start sites of both SINV22157 and SINV22107 (these two genes are in head-to-head orientation), and thus the promoters for both genes should be within this fragment (figure 2a). A change in promoters should affect gene regulation in cis and may produce ASE differences in gene expression. We examined ASE in the worker antennae RNA-seq dataset derived from SB/Sb individuals. ASE was complex for SINV23002–SINV23011, with significant higher expression of the Sb allele only at the last three sites close to the 3′ end; while for SINV22157 and SINV22107, the Sb allele was expressed significantly more than the SB allele for most SNP or indel positions (p-values less than 0.05, binomial test figure 2b and electronic supplementary material, table S5). This result indicates that the greater expression of SINV22157 in SB/Sb relative to SB/SB individuals is probably owing to the inversion swapping in a different promoter thereby causing a Sb allele-specific increase in gene expression.
4. Discussion
In this study we have revealed three aspects of the evolution of the S. invicta social supergene. First, we showed that the S. invicta supergene is composed of at least two large inversions. Second, we determined the likely inversion order for these two inversions. Last, we identified a candidate gene whose gene expression has been affected directly by the inversion breakpoint. The implications of each discovery are discussed below.
We have identified the extreme ‘right’ breakpoint for the S. invicta social supergene to nucleotide resolution, thereby establishing one-inversion boundary. This should also correspond to the supergene boundary, however because recombination is most strongly suppressed around the breakpoint [51,52] the supergene could extend a little further. Indeed the approximately 25 Kb Sb-specific insertion appears to be fixed between SB and Sb and is immediately ‘outside’ of the breakpoint. The right breakpoint is found in two invasive populations (Georgia, USA and Taiwan) indicating it is at high frequency, and possibly fixed, in the invasive range. Given that our model predicts that the extreme right breakpoint is associated with the first inversion, we propose that this same boundary would be found in the native range, and perhaps in other socially polymorphic fire ant species.
Using the right breakpoint, we were able to identify one additional large (844 Kb) inversion. Importantly, the location and orientation of an embedded unique approximately 600 bp fragment C revealed the likely order of the two major inversions. Specifically, a large (greater than 9 Mb) inversion ‘1’ occurred first, essentially capturing more than 70% of the supergene [13,27]. After, the smaller 844 Kb inversion ‘2’ occurred (which itself captured fragment C). This is the simplest model, although more complex models with additional large inversions are possible. Definitive resolution will require identification of populations or species with partial supergenes.
Previous comparisons of the SB and Sb draft genomes [13,34] revealed another small local inversion (48 Kb) [13], but not the large inversion breakpoints. The better contiguity of the PacBio Sb genome permitted identification of the additional inversion. Given our results, revisiting the previous assemblies showed that the inversion breakpoints identified in this study were at or near the scaffold ends precluding their identification.
While we have identified two large inversions, a third large inversion is predicted. Both genetic map and molecular evolution analyses indicate that the supergene extends to the left an additional approximately 1 Mb [13,27]. We could not identify the breakpoints associated with this left region. The corresponding supergene scaffolds on both the SB and Sb genomes have gaps at similar locations suggesting that the putative third inversion breakpoints are associated with scaffold ends, which further implies the inversion breakpoint probably occurred in repetitive regions. Definitive resolution of all the breakpoints will require better SB and Sb genome assemblies assisted with long reads, i.e. PacBio [32] or Oxford Nanopore [53] sequencing.
(a). Strata or lack of strata?
Large supergenes and sex chromosomes are often characterized by evolutionary strata where different segments were recruited at different times, usually attributed to the accumulation of successive inversions [54,55]. Our results revealing two large inversion fragments (and a third predicted one) would superficially suggest the potential for two (or three) strata. However, analysis of divergence in fire ants did not reveal any strata, suggesting that the fire ant supergene has only one functionally important evolutionary layer [27].
How can the molecular evolution evidence and the number of inversions be reconciled? There are at least three possible explanations. First, the large inversions could have occurred at different times, but in quick succession such that any strata would be too shallow to be detected. This possibility is supported by the young estimated age (approx. 390 000 years old) of the fire ant supergene [13]. Second, the Sb chromosome may have resulted from chromothripsis [56–58], a process first described in cancer where a chromosome shatters and then is restitched together. In this case the Sb chromosomal rearrangements were not owing to inversions per se, but were an outcome of chromosome repair. A third very interesting possibility is that the social chromosome formed in another species and then introgressed into S. invicta [24,59]. Thus, evolutionary strata would be apparent within the original species, but divergence would look uniform in S. invicta [27]. One caveat to this introgression model is that speciation must have happened long enough ago to allow divergence to accumulate to mask strata signals. Nevertheless, precedent for this model exists as the patterns of molecular divergence in a supergene in the white-throated sparrow is consistent with introgression from an extinct species [17]. Shared wing mimicry alleles in Heliconus butterflies probably also occurred via introgression [60]. Because S. invicta can hybridize with other Solenopsis species [61–64], this introgression model is possible. Future evolutionary genomic studies across more fire ant species and with more power to detect potentially shallow strata will be needed to distinguish among these scenarios.
(b). Are any of the breakpoints associated with gene mutations?
At the time of inversion formation, the gene content between the original and inversion alleles are typically identical. Given the apparent, initial, equal fitness of the two alleles, inversions can only increase in frequency through neutral drift, implying that most inversions would be lost [65,66]. However, some inversion breakpoints may fortuitously create a beneficial mutation, permitting selection to drive it to higher frequency.
We found that the gene SINV22157 was probably affected by a breakpoint mutation. This gene has higher expression in the worker antennae of SB/Sb compared to SB/SB individuals, primarily owing to Sb-allele-specific upregulation. The expression differences are probably owing to changes in the location of fragment C, which contains a putative promoter and is adjacent to the first exon of SINV22157 in SB but not in Sb (figure 2a). The separation of fragment C from SINV22157 would have occurred during the presumptive second inversion event (figure 2d). This gene has no known predicted function and may be expressed in multiple tissues because in addition to worker antennae, its expression is found in worker and queen whole-body RNA-seq datasets (electronic supplementary material, table S6).
Only a few examples are known where the inversion breakpoints affect adjacent genes [67–69]. The alteration of gene expression in SINV22157 in the fire ant social supergene system is the first description, to our knowledge, of a switch in promoters caused by an inversion breakpoint. Because of the gene expression differences, it is tempting to speculate that this mutation benefited the polygyne lifestyle and helped this new two-inversion allele replace the previous one-inversion allele. Future studies may reveal if this is the case.
5. Conclusion
In summary, this study advances our understanding of the evolution of the fire ant supergene. We have identified the right-most extreme breakpoint for the supergene. Aided by this breakpoint, comparisons between the genome assemblies revealed an additional large inversion (844 Kb) towards the left end of the supergene. Furthermore, the translocation of a unique approximately 600 bp fragment revealed the most probable historical order of these two inversions. While the supergene did not appear to exhibit evolutionary strata in S. invicta, future evolutionary genomic studies across more fire ant species will ultimately clarify if these inversions may correspond to evolutionary strata elsewhere. Finally, we have identified a rare example of gene expression alteration caused directly by the breakpoint mutation. It remains to be determined if this mutation was beneficial for the polygyne lifestyle in fire ants and will be the subject of future studies.
Supplementary Material
Acknowledgements
We thank Wen-Hsiung Li and three reviewers for valuable insights and comments on the manuscript; the Taiwan National Center for High-performance Computing for computer time and facilities; Meiyeh Lu and the High Throughput Sequencing Core hosted in the Biodiversity Research Center in Academia Sinica for sequencing, advice and support. Research was supported by the Biodiversity Research Center, Academia Sinica, grants from MOST (100–2311-B-001-015-MY3, 103-2311-B-001-018-MY3, 104-2314-B-001-009-MY5), and an Academia Sinica Career Development Grant to J.W.
Data accessibility
GeneBank accession numbers of DNA sequences: six S. invicta Sb contigs (MH121689 to MH121694), one S. geminata contig (MH121695), and the end sequences of six BACs (MH121696 to MH121701). Data and analyses are deposited in Dryad at (http://dx.doi.org/10.5061/dryad.2458p4r) [28].
Authors' contributions
Y-C.H., V.D.D. and J.W. conceived and designed the experiments. N-C.C. initiated this project. Y-C.H., V.D.D. and N-C.C. conducted molecular biology experiments. Y-C.H. conducted cytological experiments. Y-C.H., V.D.D. and J.W. carried out bioinformatic analyses. Y-C.H. contributed to all stages of the project. Y-C.H., V.D.D. and J.W. wrote the paper. All authors gave final approval for publication.
Competing interests
We declare we have no competing interests.
References
- 1.Thompson MJ, Jiggins CD. 2014. Supergenes and their role in evolution. Heredity 113, 1–8. ( 10.1038/hdy.2014.20) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schwander T, Libbrecht R, Keller L. 2014. Supergenes and complex phenotypes. Curr. Biol. 24, R288–R294. ( 10.1016/j.cub.2014.01.056) [DOI] [PubMed] [Google Scholar]
- 3.Dobzhansky T. 1947. Adaptive changes induced by natural selection in wild populations of Drosophila. Evolution Int. J. Org Evolution 1, 1–16. ( 10.1111/j.1558-5646.1947.tb02709.x) [DOI] [Google Scholar]
- 4.Dobzhansky T. 1970. Genetics of the evolutionary process. New York, NY: Columbia University Press. [Google Scholar]
- 5.Clarke CA, Sheppard PM. 1971. Further studies on genetics of mimetic butterfly Papilio-Memnon L. Phil. Trans. R. Soc. Lond. B 263, 37–70. ( 10.1098/rstb.1971.0109) [DOI] [PubMed] [Google Scholar]
- 6.Pamela V, Dowrick J. 1956. Heterostyly and homostyly in Primula obconica. Heredity 10, 219–236. ( 10.1038/hdy.1956.19) [DOI] [Google Scholar]
- 7.Mather K. 1950. The genetical architecture of heterostyly in Primula-Sinensis. Evol. Int. J. Org Evol. 4, 340–352. ( 10.2307/2405601) [DOI] [Google Scholar]
- 8.Lyttle TW. 1993. Cheaters sometimes prosper—distortion of Mendelian segregation by meiotic drive. Trends Genet. 9, 205–210. ( 10.1016/0168-9525(93)90120-7) [DOI] [PubMed] [Google Scholar]
- 9.Larracuente AM, Presgraves DC. 2012. The selfish segregation distorter gene complex of Drosophila melanogaster. Genetics 192, 33–53. ( 10.1534/genetics.112.141390) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lyon MF. 2003. Transmission ratio distortion in mice. Annu. Rev. Genet. 37, 393–408. ( 10.1146/annurev.genet.37.110801.143030) [DOI] [PubMed] [Google Scholar]
- 11.Dobrovolskaia-Zavadskaia N. 1927. Regarding the spontaneous mortification of the tail of a new-born mouse and the existence of a hereditary characteristic (factor). C. R. Soc. Biol. 97, 114–116. [Google Scholar]
- 12.Hiraizumi Y, Crow JF. 1957. The amount of dominance of ‘recessive’ lethals from natural populations of D. melanogaster. Drosophila Inform. Serv. 31, 123. [Google Scholar]
- 13.Wang J, Wurm Y, Nipitwattanaphon M, Riba-Grognuz O, Huang YC, Shoemaker D, Keller L. 2013. A Y-like social chromosome causes alternative colony organization in fire ants. Nature 493, 664–668. ( 10.1038/Nature11832) [DOI] [PubMed] [Google Scholar]
- 14.Purcell J, Brelsford A, Wurm Y, Perrin N, Chapuisat M. 2014. Convergent genetic architecture underlies social organization in ants. Curr. Biol. 24, 2728–2732. ( 10.1016/j.cub.2014.09.071) [DOI] [PubMed] [Google Scholar]
- 15.Kupper C, et al. 2016. A supergene determines highly divergent male reproductive morphs in the ruff. Nat. Genet. 48, 79–83. ( 10.1038/ng.3443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamichhaney S, et al. 2016. Structural genomic changes underlie alternative reproductive strategies in the ruff (Philomachus pugnax). Nat. Genet. 48, 84–88. ( 10.1038/ng.3430) [DOI] [PubMed] [Google Scholar]
- 17.Tuttle EM, et al. 2016. Divergence and functional degradation of a sex chromosome-like supergene. Curr. Biol. 26, 344–350. ( 10.1016/j.cub.2015.11.069) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Joron M, et al. 2011. Chromosomal rearrangements maintain a polymorphic supergene controlling butterfly mimicry. Nature 477, 203–206. ( 10.1038/nature10341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kunte K, Zhang W, Tenger-Trolander A, Palmer DH, Martin A, Reed RD, Mullen SP, Kronforst MR. 2014. Doublesex is a mimicry supergene. Nature 507, 229–232. ( 10.1038/nature13112) [DOI] [PubMed] [Google Scholar]
- 20.Roberts RB, Ser JR, Kocher TD. 2009. Sexual conflict resolved by invasion of a novel sex determiner in Lake Malawi cichlid fishes. Science 326, 998–1001. ( 10.1126/science.1174705) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lowry DB, Willis JH. 2010. A widespread chromosomal inversion polymorphism contributes to a major life-history transition, local adaptation, and reproductive isolation. PLoS Biol. 8, e1000500 ( 10.1371/journal.pbio.1000500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keller L, Ross KG. 1998. Selfish genes: a green beard in the red fire ant. Nature 394, 573–575. ( 10.1038/29064) [DOI] [Google Scholar]
- 23.Huang YC, Wang J. 2014. Did the fire ant supergene evolve selfishly or socially? BioEssays 36, 200–208. ( 10.1002/bies.201300103) [DOI] [PubMed] [Google Scholar]
- 24.Gotzek D, Ross KG. 2007. Genetic regulation of colony social organization in fire ants: an integrative overview. Q Rev. Biol. 82, 201–226. ( 10.1086/519965) [DOI] [PubMed] [Google Scholar]
- 25.Krieger MJ, Ross KG. 2002. Identification of a major gene regulating complex social behavior. Science 295, 328–332. ( 10.1126/science.1065247) [DOI] [PubMed] [Google Scholar]
- 26.Ross KG. 1997. Multilocus evolution in fire ants: effects of selection, gene flow and recombination. Genetics 145, 961–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pracana R, Priyam A, Levantis I, Nichols RA, Wurm Y. 2017. The fire ant social chromosome supergene variant Sb shows low diversity but high divergence from SB. Mol. Ecol. 26, 2864–2879. ( 10.1111/mec.14054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang YC, Dang VD, Chang NC, Wang J.. 2018. Data from: Multiple large inversions and breakpoint rewiring of gene expression in the evolution of the fire ant social supergene Dryad Digital Repository. ( 10.5061/dryad.2458p4r) [DOI] [PMC free article] [PubMed]
- 29.Huang YC, Lee CC, Kao CY, Chang NC, Lin CC, Shoemaker D, Wang J. 2016. Evolution of long centromeres in fire ants. BMC Evol. Biol. 16, ARTN 189 ( 10.1186/s12862-016-0760-7) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Imai HT. 2016. A manual for ant chromosome preparations (an improved air-drying method) and Giemsa staining. Chromosome Sci. 19, 57–66. [Google Scholar]
- 31.Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, Rozen SG. 2012. Primer3-new capabilities and interfaces. Nucleic Acids Res. 40, ARTN e115 ( 10.1093/nar/gks596) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rhoads A, Au KF. 2015. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics 13, 278–289. ( 10.1016/j.gpb.2015.08.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carver TJ, Rutherford KM, Berriman M, Rajandream MA, Barrell BG, Parkhill J. 2005. ACT: the Artemis comparison tool. Bioinformatics 21, 3422–3423. ( 10.1093/bioinformatics/bti553) [DOI] [PubMed] [Google Scholar]
- 34.Wurm Y, et al. 2011. The genome of the fire ant Solenopsis invicta. Proc. Natl Acad. Sci. USA 108, 5679–5684. ( 10.1073/pnas.1009690108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chaisson MJ, Tesler G. 2012. Mapping single molecule sequencing reads using basic local alignment with successive refinement (BLASR): application and theory. BMC Bioinformatics 13, Artn 238 ( 10.1186/1471-2105-13-238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26. ( 10.1038/nbt.1754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Pyl PT, Huber W. 2015. HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. ( 10.1093/bioinformatics/btu638) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140. ( 10.1093/bioinformatics/btp616) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKenna A, et al. 2010. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.DePristo MA, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. ( 10.1038/ng.806) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van der Auwera GA, et al. 2013. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33. ( 10.1002/0471250953.bi1110s43) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruitt KD, et al. 2014. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 42, D756–D763. ( 10.1093/nar/gkt1114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. ( 10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 45.Caceres M, Ranz JM, Barbadilla A, Long M, Ruiz A. 1999. Generation of a widespread Drosophila inversion by a transposable element. Science 285, 415–418. ( 10.1126/science.285.5426.415) [DOI] [PubMed] [Google Scholar]
- 46.Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu. Rev. Genet. 41, 331–368. ( 10.1146/annurev.genet.40.110405.090448) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Casals F, Caceres M, Ruiz A. 2003. The foldback-like transposon Galileo is involved in the generation of two different natural chromosomal inversions of Drosophila buzzatii. Mol. Biol. Evol. 20, 674–685. ( 10.1093/molbev/msg070) [DOI] [PubMed] [Google Scholar]
- 48.Zhang JB, Peterson T. 2004. Transposition of reversed Ac element ends generates chromosome rearrangements in maize. Genetics 167, 1929–1937. ( 10.1534/genetics.103.026229) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gray YHM. 2000. It takes two transposons to tango: transposable-element-mediated chromosomal rearrangements. Trends Genet. 16, 461–468. ( 10.1016/S0168-9525(00)02104-1) [DOI] [PubMed] [Google Scholar]
- 50.Puig M, Casillas S, Villatoro S, Caceres M. 2015. Human inversions and their functional consequences. Brief Funct. Genomics 14, 369–379. ( 10.1093/bfgp/elv020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navarro A, Bardadilla A, Ruiz A. 2000. Effect of inversion polymorphism on the neutral nucleotide variability of linked chromosomal regions in Drosophila. Genetics 155, 685–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andolfatto P, Depaulis F, Navarro A. 2001. Inversion polymorphisms and nucleotide variability in Drosophila. Genet. Res. 77, 1–8. ( 10.1017/S0016672301004955) [DOI] [PubMed] [Google Scholar]
- 53.Laver T, Harrison J, O'Neill PA, Moore K, Farbos A, Paszkiewicz K, Studholme DJ. 2015. Assessing the performance of the Oxford Nanopore Technologies MinION. Biomol. Detect. Quantif. 3, 1–8. ( 10.1016/j.bdq.2015.02.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 14, 113–124. ( 10.1038/nrg3366) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Charlesworth D, Charlesworth B, Marais G. 2005. Steps in the evolution of heteromorphic sex chromosomes. Heredity 95, 118–128. ( 10.1038/sj.hdy.6800697) [DOI] [PubMed] [Google Scholar]
- 56.Stephens PJ, et al. 2011. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40. ( 10.1016/j.cell.2010.11.055) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Forment JV, Kaidi A, Jackson SP. 2012. Chromothripsis and cancer: causes and consequences of chromosome shattering. Nat. Rev. Cancer 12, 663–670. ( 10.1038/nrc3352) [DOI] [PubMed] [Google Scholar]
- 58.Maher CA, Wilson RK. 2012. Chromothripsis and human disease: piecing together the shattering process. Cell 148, 29–32. ( 10.1016/j.cell.2012.01.006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keller L., Parker J.D. 2002. Behavioral genetics: a gene for supersociality. Curr. Biol. 12, R180–R181. ( 10.1016/S0960-9822(02)00737-6) [DOI] [PubMed] [Google Scholar]
- 60.Smith J, Kronforst MR. 2013. Do Heliconius butterfly species exchange mimicry alleles? Biol. Lett. 9, 20130503 ( 10.1098/rsbl.2013.0503) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shoemaker DD, Ahrens ME, Ross KG. 2006. Molecular phylogeny of fire ants of the Solenopsis saevissima species-group based on mtDNA sequences. Mol. Phylogenet. Evol. 38, 200–215. ( 10.1016/j.ympev.2005.07.014) [DOI] [PubMed] [Google Scholar]
- 62.Ross KG, Shoemaker DD. 2005. Species delimitation in native South American fire ants. Mol. Ecol. 14, 3419–3438. ( 10.1111/j.1365-294X.2005.02661.x) [DOI] [PubMed] [Google Scholar]
- 63.Shoemaker DD, Ross KG, Arnold ML. 1996. Genetic structure and evolution of a fire ant hybrid zone. Evol. Int. J. Org. Evol. 50, 1958–1976. ( 10.1111/j.1558-5646.1996.tb03583.x) [DOI] [PubMed] [Google Scholar]
- 64.Ross KG, Robertson JL. 1990. Developmental stability, heterozygosity, and fitness in 2 introduced fire ants (Solenopsis invicta and Solenopsis richteri) and their hybrid. Heredity 64, 93–103. ( 10.1038/hdy.1990.12) [DOI] [Google Scholar]
- 65.Kirkpatrick M. 2010. How and why chromosome inversions evolve. PLoS Biol. 8, ARTN e1000501 ( 10.1371/journal.pbio.1000501) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hoffmann AA, Rieseberg LH. 2008. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu. Rev. Ecol. Evol. Syst. 39, 21–42. ( 10.1146/annurev.ecolsys.39.110707.173532) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Castermans D, Vermeesch JR, Fryns JP, Steyaert JG, Van de Ven WJ, Creemers JW, Devriendt K. 2007. Identification and characterization of the TRIP8 and REEP3 genes on chromosome 10q21.3 as novel candidate genes for autism. Eur. J. Hum. Genet. 15, 422–431. ( 10.1038/sj.ejhg.5201785) [DOI] [PubMed] [Google Scholar]
- 68.Puig M, Caceres M, Ruiz A. 2004. Silencing of a gene adjacent to the breakpoint of a widespread Drosophila inversion by a transposon-induced antisense RNA. Proc. Natl Acad. Sci. USA 101, 9013–9018. ( 10.1073/pnas.0403090101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Edwards PA. 2010. Fusion genes and chromosome translocations in the common epithelial cancers. J. Pathol. 220, 244–254. ( 10.1002/path.2632) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Huang YC, Dang VD, Chang NC, Wang J.. 2018. Data from: Multiple large inversions and breakpoint rewiring of gene expression in the evolution of the fire ant social supergene Dryad Digital Repository. ( 10.5061/dryad.2458p4r) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
GeneBank accession numbers of DNA sequences: six S. invicta Sb contigs (MH121689 to MH121694), one S. geminata contig (MH121695), and the end sequences of six BACs (MH121696 to MH121701). Data and analyses are deposited in Dryad at (http://dx.doi.org/10.5061/dryad.2458p4r) [28].