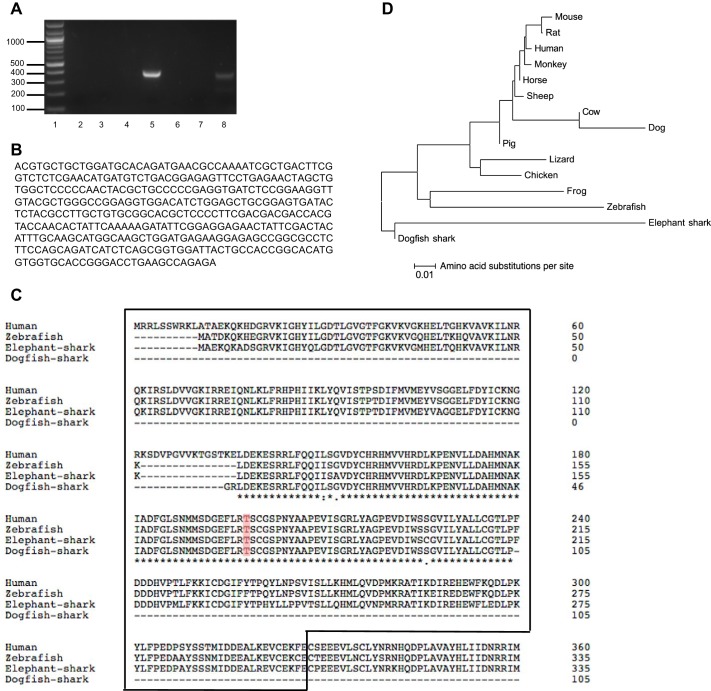
Fig. 2.
Cloning of a 389-bp DNA sequence of the AMPKα subunit in the dogfish shark rectal gland. A: PCR of shark rectal gland cDNA yielded a 389-bp product of AMPKα1 using degenerate primers designed from conserved regions of human, cow, rat, pig, chicken, and frog AMPKα1 (lane 5). Shown are a 100-bp DNA ladder (lane 1), PCR with different pairs of AMPKα1 degenerate primers against regions of the gene (lanes 2–7), and a positive control using TASK-1 potassium channel degenerate primers (394-bp PCR product, lane 8). B: cloned 389-bp DNA sequence of the catalytic kinase domain of the AMPKα subunit in the shark rectal gland. C: amino acid alignment of the dogfish shark fragment and of AMPKα from human, zebrafish, and elephant shark. Asterisk (*) indicates the amino acids are identical; colon (:) indicates that conserved substitutions are observed; and period (.) indicates that semiconserved substitutions are observed. The catalytic domain of the protein is enclosed in a box. The phosphorylation residue Thr172 is marked in color. D: phylogenetic tree showing the relationship of vertebrate and invertebrate AMPKα. Protein alignments were performed with ClustalW. The evolutionary history was inferred using the Neighbor-Joining method. The optimal tree with a sum of branch length of 0.36877630 is shown. The tree is drawn to scale 0.01, with branch lengths and evolutionary distances both in amino acid substitutions per site. The evolutionary distances were computed using the Poisson correction method. Rate variation among sites was modeled with a gamma distribution (shape parameter = 1). The analysis involved 15 amino acid sequences. All ambiguous positions were removed for each sequence pair. There were a total of 620 positions in the final data set. Evolutionary analyses were conducted in MEGA6.