Figure 1.
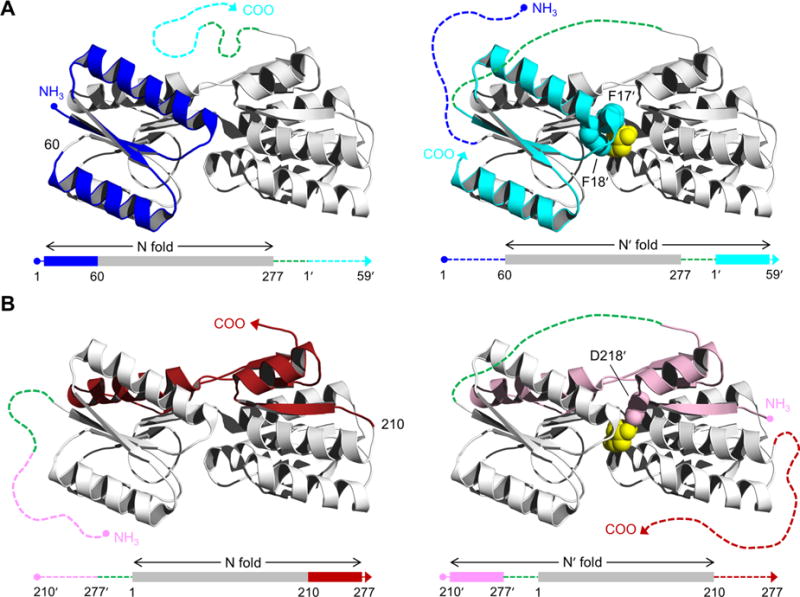
Structure of RBP and schematic of the AFF design. RBP binds ribose (yellow spheres) in the cleft between amino- and carboxy-terminal domains. AFF variants are generated by duplicating either (A) an amino-terminal segment (blue) and fusing it to the carboxy terminus (cyan), as exemplified by AFF60, or (B) a carboxy-terminal segment (red) and appending it to the amino terminus (pink), as demonstrated by AFF210. The N and N′ conformations are shown at THE left and right, respectively. In the amino acid sequences, depicted below each structure, rectangles indicate folded structure and dashed lines represent unfolded/unstable structure. The dashed green line is a 30-amino acid linker. The copy of the duplicated segment that is orphaned in each conformation is depicted as a dashed line extending from one of the termini. Critical ribose contacts are established by Phe17 and Phe18 (cyan spheres in panel A) and by Asp218 (pink spheres in panel B). Substrate binding to the N conformations of AFF60 and AFF210 is abolished by introducing the F17A/F18A double mutation and the D218S mutation into the blue and red regions, respectively. Addition of ribose, and the binding energy provided by Phe17′ and Phe18′ (AFF60) or Asp218′ (AFF210) in the remodeled binding pocket, drive the shift from the N fold to the N′ fold. The folded blue segment of AFF60 becomes an amino-terminal tail, and the cyan carboxy-terminal tail becomes part of the folded protein. The analogous event happens with the red and pink regions of AFF210 except the locations of the tails are reversed. To detect this rearrangement, fluorophores are placed at the site of permutation (position 60 in AFF60, position 210 in AFF210, etc.) and at the carboxy termini of AFF60 and AFF70 or the amino termini of AFF186 and AFF210.
