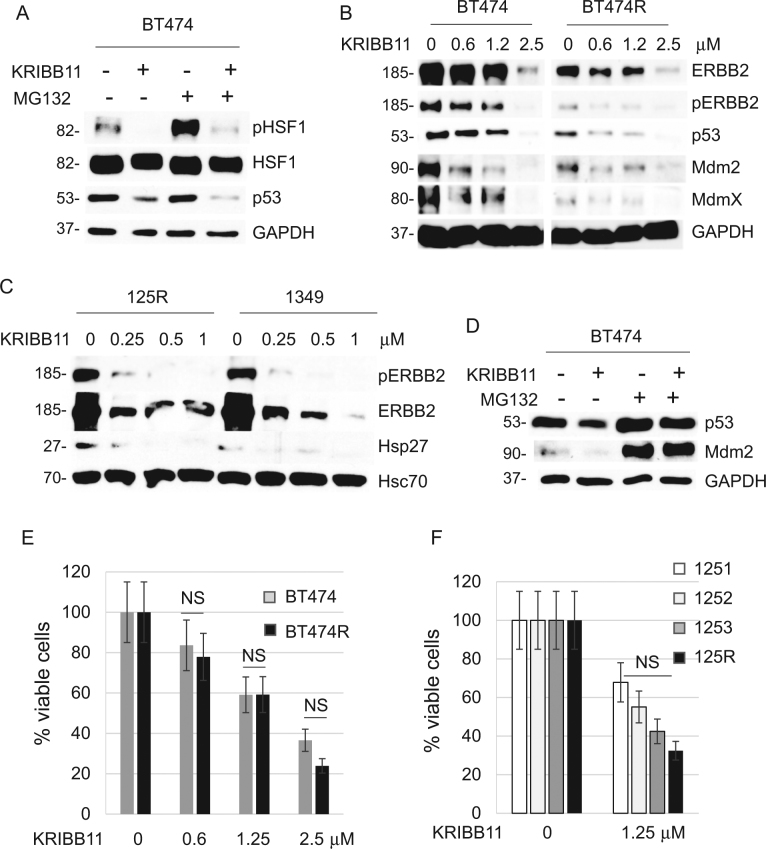
Fig. 5. HSF1 inhibition causes degradation of mutp53 and ErbB2, and suppresses growth of both lapatinib-sensitive and lapatinib-resistant cancer cells.
a–d The HSF1 inhibitor KRIBB11 suppresses activation of HSF1 (measured by phospho-Ser326) after MG132-induced proteotoxic stress (1 μM, 2.5 h) in lapatinib-sensitive BT474 cells (a), suppresses ERBB2 signaling and destabilizes mutp53 in both lapatinib-sensitive BT474 and lapatinib-resistant BT474R cells (b), suppresses ERBB2 signaling and HSF1 target Hsp27 in both lapatinib-sensitive 1349 and lapatinib-resistant 125R murine cells (c); and induces degradation of mutp53 and Mdm2 in lapatinib-sensitive BT474 cells, which is rescued by the proteasome inhibitor MG132 (d). Cells were pre-treated with KRIBB11 (2.5 μM, 24 h) followed by MG132 treatment (1 μM, 2.5 h) (a) or simultaneously treated with KRIBB11 (2.5 μM) and MG132 (2.5 μM) for 24 h (d). Western blot analyses, GAPDH and Hsc70 served as a loading control. e, f The HSF1 inhibitor KRIBB11 suppresses growth of human BT474R (e) and mouse 125R (f) lapatinib-resistant cells as efficiently as their corresponding lapatinib-sensitive controls, BT474 and 1251, 1252, 1349 cells, respectively. Cells were treated with indicated concentrations of KRIBB11 for 48 h, followed by cell viability assays, which are shown relative to DMSO-treated cells. One representative experiment out of two independent experiments (each performed in triplicate) is shown; NS non-significant