Figure 1. IFNGR1 is ubiquitinated and degraded by the proteasome.
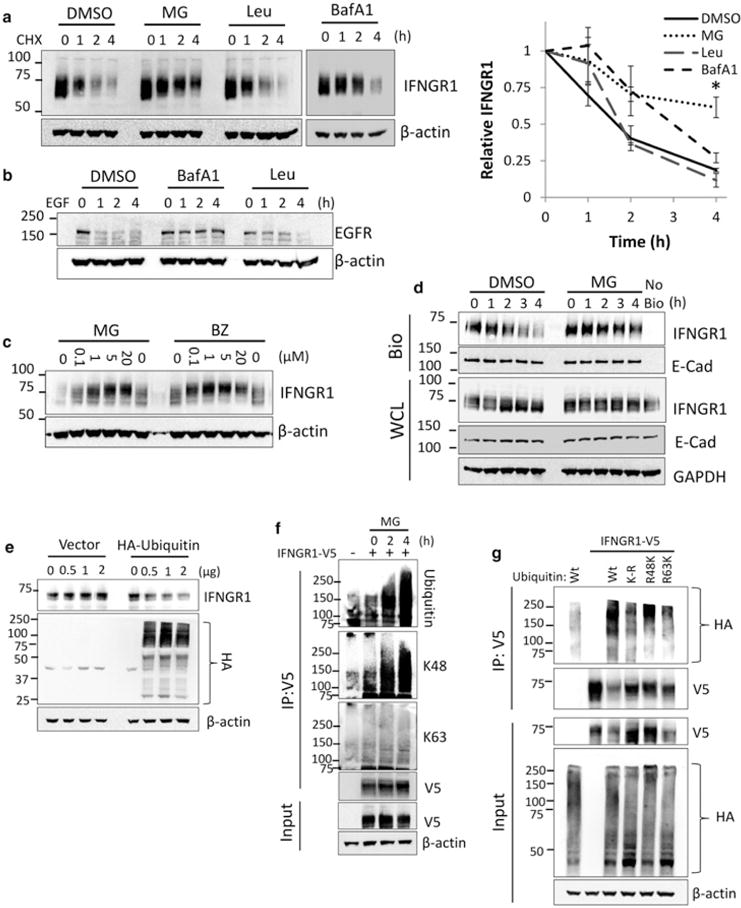
(a) A549 cells were treated with CHX for the indicated times in the presence of DMSO, MG132 (10 μM), bafilomycin A1 (BafA1) (100 nM) or leupeptin (Leu) (100 μM). Cells were then harvested and probed for IFNGR1 expression. Right: IFNGR1 protein densitometry normalized to β-actin and CHX 0 h to determine protein half-life. n = 3–4 independent experiments, ±SE. *P < 0.05 vs. control by ANOVA and Tukey’s t-test. (b) A549 cells were treated with EGF (100 μM) for the indicated times in the presence of DMSO, BafA1, or Leu at the concentrations above. Cells were then harvested and probed for EGFR expression. (c) A549 cells were treated with indicated concentrations of MG132 or bortezomib (BZ) for 4 h, harvested, and probed for IFNGR1 expression. (d) A549 cells were incubated with membrane impermeable biotin at 4°C, returned to the incubator and treated with DMSO or MG132 for indicated times. Cells were probed for plasma membrane (biotinylated, [Bio]) and whole cellular lysates (WCL) IFNGR1. E-cadherin was included as a plasma membrane marker, no bio = no biotinylation control. n = 2 independent experiments (e) HEK cells were transfected with HA-ubiquitin or an empty vector control. At 24 h, cells were harvested and probed for endogenous IFNGR1. The middle blot below shows levels of total cellular ubiquitination. (f) HEK cells were transfected with V5-tagged IFNGR1. At 2 and 4 h prior to harvest cells were treated with 10 μM MG132. 48 h post-transfection cell lysates were boiled, immunoprecipitated (IP) for V5, and probed for total ubiquitin, K48-linked ubiquitin, and K63-linked ubiquitin; n = 3 independent experiments. (g) HEK cells were transfected with V5-tagged IFNGR1 and Wt, lysine-less (K-R), lysine 48 only (R48K), and lysine 63 only (R63K) HA-tagged ubiquitin plasmids. At 4 h prior to harvest cells were treated with 10 μM MG132. 48 h post-transfection cell lysates were boiled, subjected to IP for V5, and probed for HA; n = 2 independent experiments.
