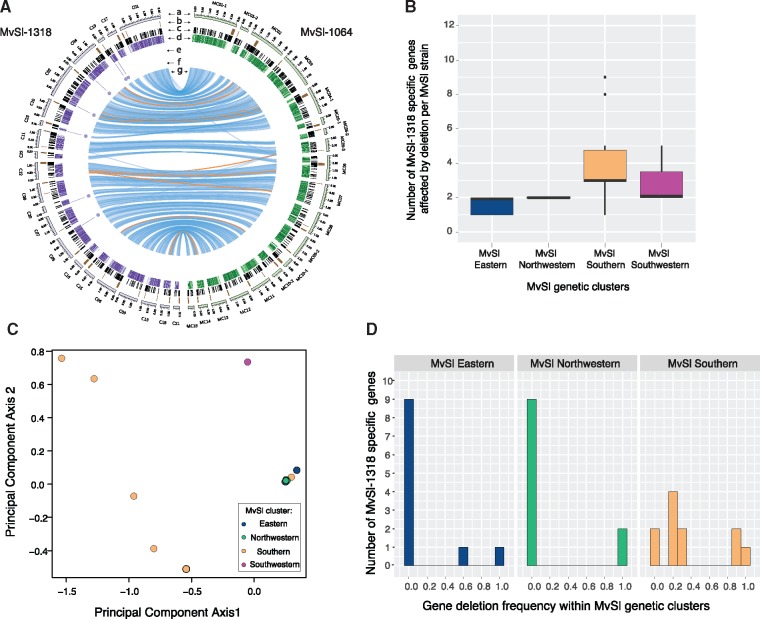
Fig. 6.
—Identification of strain-specific genes in three Microbotryum lychnidis-dioicae (MvSl) de novo assembled genomes and effect of population structure on gene presence–absence polymorphism. (A) Comparison between high-quality genome assemblies of the MvSl strains MvSl-1064 and MvSl-1318. The different tracks are: (a) Contigs of the MvSl-1318 (left, purple tracks) and MvSl-1064 reference genomes (right, green tracks). Only contigs >320 kb were represented; (b) Location of centromeric repeats; (c) Location of transposable elements; (d) Gene density in 10-kb nonoverlapping windows (the gradient shows differences from 0% to 100%); (e) Location of detected missing fragments in MvSl-1064; (f) Location of missing genes in MvSl-1064, that is, MvSl-1318-specific genes; (g) Links representing collinearity of genomic regions >10 kb between the MvSl-1064 and MvSl-1318 reference genomes, with the orange links corresponding to inversions. (B) Distribution of the number of MvSl-1318-specific genes predicted to be absent per MvSl strain and per genetic cluster. (C) Principal component analysis of gene presence–absence polymorphism of MvSl-1318-specific genes in MvSl strains based on six unique gene presence–absence polymorphisms, that is, using only a single gene per missing fragments. Points correspond to strains and are colored according to the genetic clusters identified by the dendrogram (fig. 4D). (D) Distribution of gene absence frequency of MvSl-1318-specific genes in the four MvSl genetic clusters.