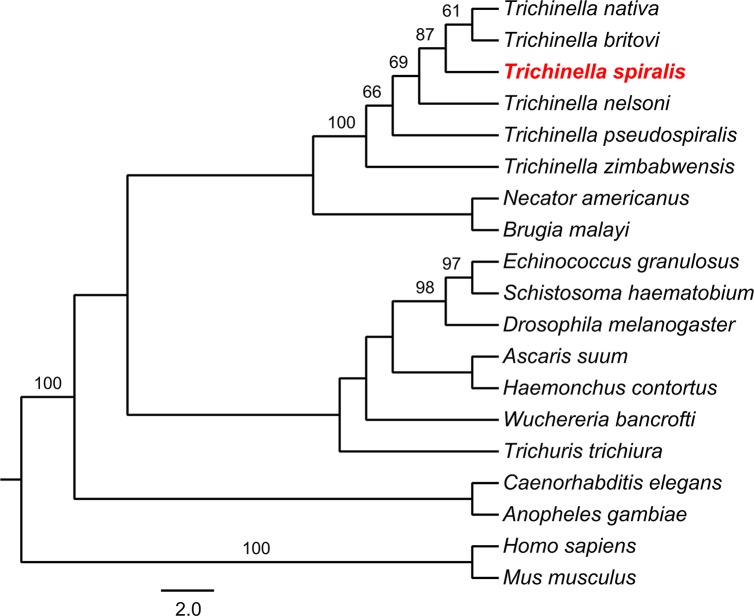
Fig 3. Cladogram of analysis of TsSP.
The maximum parsimony tree of serine protease from 19 organisms calculated using MEGA. The GenBank accession numbers of each serine protease are as follows: Trichinella spiralis (ABY60762.1), Trichinella nativa (KRZ48330.1), Trichinella nelsoni (KRX27556.1), Trichinella britovi (KRY59723.1), Trichinella pseudospiralis (KRY01512.1), Trichinella zimbabwensis (KRZ02345.1), Homo sapiens (CAB91984.1), Mus musculus (EDL11329.1), Caenorhabditis elegans (CCD65324.1), Drosophila melanogaster (NP_001262565.1), Trichuris trichiura (CDW53654.1), Ascaris suum (ERG81033.1), Necator americanus (XP_013308358.1), Brugia malayi (XP_001900088.1), Wuchereria bancrofti (EJW79555.1), Haemonchus contortus (CDJ82043.1), Echinococcus granulosus (EUB62466.1), Anopheles gambiae (CAB90819.1), Schistosoma haematobium (KGB34910.1). Bootstrap values which are higher than 80 are indicated on branches.