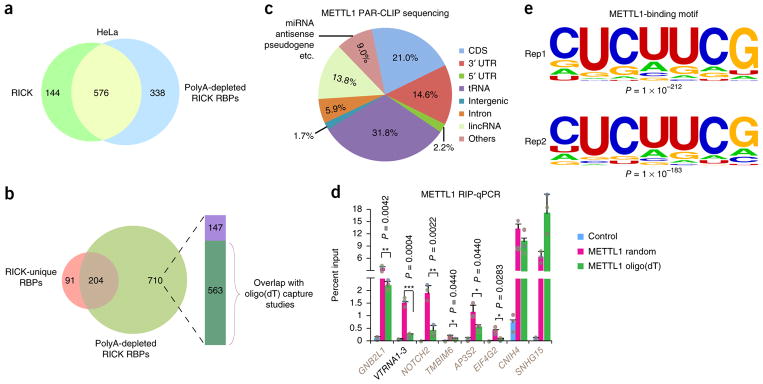
Figure 5.
Identification of METTL1-interacting RNAs. (a) Venn diagram comparing the 720 high-confidence proteins identified by RICK and the 914 high-confidence proteins identified by polyA-depleted RICK. (b) Venn diagram comparing the 295 RICK-unique RBPs and the 914 high-confidence proteins identified by polyA-depleted RICK. The column indicates distribution of the 710 polyA-depleted RICK proteins compared to high-confidence proteins of different oligo(dT) capture studies (green, proteins present in different oligo(dT) capture data sets1,2,18,19; purple, proteins not present in the same oligo(dT) capture data sets). (c) Distribution of RNA species identified by METTL1 PAR-CLIP sequencing. The average of n = 2 biologically independent experiments is shown. (d) RIP-qPCR analysis of different RNAs binding to METTL1 that were identified in the METTL1 PAR-CLIP sequencing. The enrichments were normalized to input; GFP was used as a negative control. Random hexamers or oligo(dT) primers were used for the reverse transcription as indicated. METTL1-interacting mRNAs are marked in gray as opposed to VTRNA1-3; n = 3 biologically independent experiments, and data are shown as the mean ± s.d.; P value is shown (Student’s t-test, two tailed). (e) The most enriched METTL1-binding motifs on all target RNAs (n = 26,311 and 20,816 for Rep1 and Rep2, respectively) identified in two independent PAR-CLIP sequencing experiments; P value is shown (Fisher’s exact test, one tailed).