Figure 2.
Characterization of the Effects of the ID-Associated Mutation on OGT TPR Stability and Dynamics
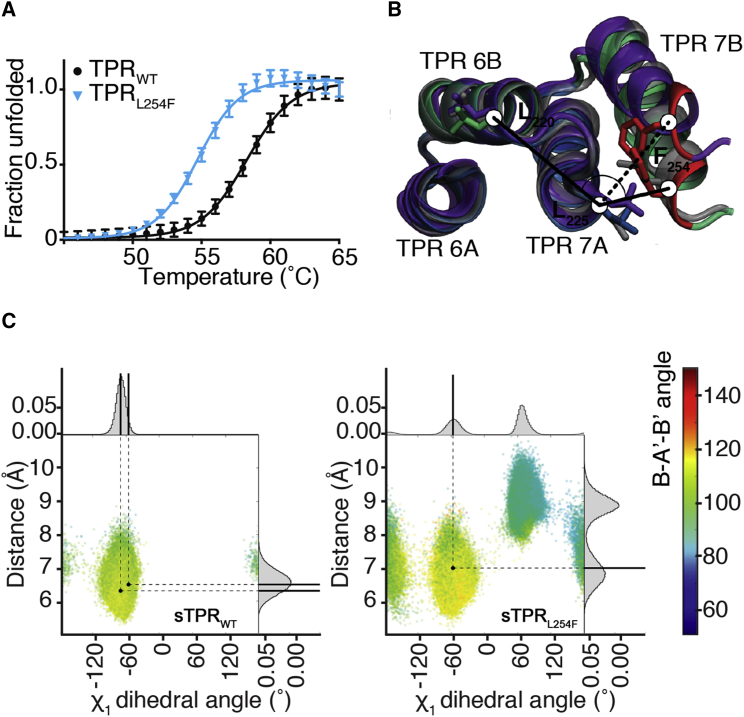
(A) Thermal denaturing curve showing fraction of unfolded TPRWT and TPRL254F constructs as a function of temperature. Data averaged from seven replicates were fitted to a Boltzmann sigmoidal curve equation, with error bars representing SD.
(B) Superposition of sTPRWT (gray), sTPRL254F-C1 (green), and sTPRL254F-C2 (purple), with the B-A′-B′ angle and intra-TPR distance demarcated with solid and dashed lines, respectively. See also Figures S2 and S3.
(C) Graphs of sTPRWT (left) and sTPRL254F (right) conformational populations in the molecular dynamics simulations, with the χ1 dihedral angle of residue 254 shown on the x axis, the intra-TPR repeat distance shown on the y axis, and the angle B-A′-B′ shown as a color scale. The B-A′-B′ values observed in the crystal structures are shown as black dots. Histograms attached to the graph show the distribution of χ1 dihedral angles. See also Figures S2 and S3.
sTPRL254F-C1 and sTPRL254F-C1, sTPRL254F conformations 1 and 2; sTPR, simplified TPR; TPR, tetratricopeptide repeats.