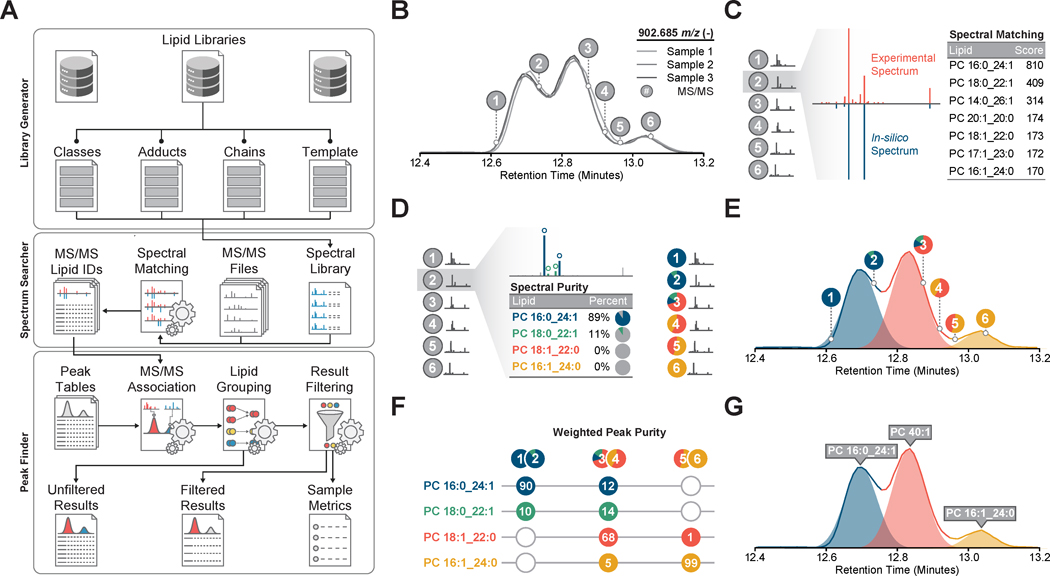
Figure 1. Lipid Identification and Quantitation Workflow.
(A) Overview of library construction (Library Generator), lipid identification (Spectrum Searcher), and result filtering (Peak Finder) in LipiDex.
(B) Extraction and alignment of lipid chromatographic peaks using external software packages.
(C) Identification of MS/MS spectra (grey circles and red spectrum) using in-silico lipid databases and a modified dot-product scoring algorithm which ranks identifications according to spectral similarity to library entries (blue spectrum).
(D) Spectral deconvolution and relative quantitation (colored circles) of co-fragmented isobaric lipid species by sequential matching against and subtracting from the experimental spectra.
(E) Accurate association of MS/MS identifications to chromatographic features via Gaussian peak modeling (shaded color).
(F) Calculation of total peak purity for each chromatographic peak across all samples using the Gaussian peak profile to modify spectral purity measurements.
(G) Final identification of each chromatographic peak using either the sum composition (i.e. PC40:1) or molecular composition (i.e. PC 16:1_24:0) if the weighted peak purity rises above a user-supplied threshold.