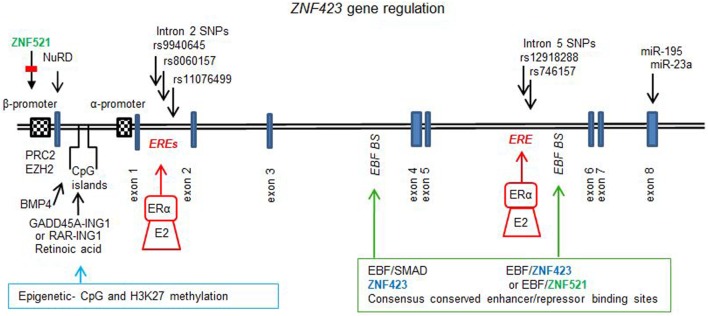
Figure 1.
ZNF423 gene regulation. Diagram to illustrate the structure for the ZNF423 human (NG_032972.2, 377, 389 bp) gene, which comprises 8 exons and introns with the major part of the protein being coded by exon 4. Two alternative promoters have been described (21, 59), and these are subject to regulation by demethylation and methylation of the CpG islands controlled by GADD45-inhibitor of growth protein 1 (ING1) or retinoic acid induced RAR–ING1 complexes (68). Epigenetic reprogramming is also associated with histone methylation (H3K9me3) and acetylation (H3K9ac), controlled by PCR2 and EHZ2 (56) as well as by ZNF521 (33). BMP4 can result in the demethylation of specific CpG dinucleotides resulting in activation of transcription (40). Consensus conserved across species sites for ZNF423, early B cell factor (EBF), and small mother against decapentaplegic have been found in introns 3 and 5. The enhancer site in intron 5 was found to be occupied by Zfp423 or Zfp521 together with EBF1 resulting in autoregulation by Zfp423 (34) or repression by Zfp521 (33). In ER+ breast cancers, single nucleotide polymorphisms in intron 2 (12) and in intron 5 (15) have been found, which are sufficiently close to ERα canonical consensus-binding sites [estrogen response element (ERE)] such that the response to E2 and selective estrogen receptor modulators is affected. Additionally, the 3′UTR of the ZNF423 mRNA contains sites for miR195 (35, 53) and miR23a (36), which control expression.