Abstract
A diverse spectrum of intracellular bacterial pathogens that inhabit the cytosol have evolved the ability to polymerize actin on their surface to power intracellular actin-based motility (ABM). These include species of Listeria, Burkholderia and Rickettsia, as well as Shigella and Mycobacteria. Here, we provide an overview of the roles of bacterial ABM in survival and virulence. Moreover, we survey the molecular mechanisms of actin polymerization in host cells and describe how bacterial pathogens mimic or harness the full diversity of these mechanisms for ABM. Finally, we present ABM through a new lens by comparing motility mechanisms between related species of Listeria, Burkholderia and Rickettsia. Through these comparisons, we hope to illuminate how exploitation of different actin polymerization mechanisms influences ABM as well as pathogenicity and virulence in humans and other animals.
Keywords: bacterial pathogen, Listeria, Burkholderia, Rickettsia, cytoskeleton, actin-based motility
This minireview explores the roles of bacterial pathogen actin-based motility in survival and cell–cell spread, the diverse spectrum of motility mechanisms, and the impact of this mechanistic diversity on pathogenicity and virulence.
INTRODUCTION
Intracellular bacterial pathogens remodel and exploit the host cell environment to support their survival and growth. A common target of bacterial pathogens is the host cell actin cytoskeleton, which is a dynamic system of filaments that is central to shape determination, movement, phagocytosis and intracellular trafficking. Because of its importance in these processes, the actin cytoskeleton is manipulated by many bacterial pathogens at multiple stages of infection (Fig. 1) (reviewed in Haglund and Welch 2011). For example, most intracellular bacterial pathogens mobilize actin during invasion (reviewed in Carabeo 2011). Following invasion, some pathogens remain within a membrane-bound compartment and target actin to subvert membrane trafficking (Haglund and Welch 2011). Perhaps the most striking mobilization occurs when bacteria that escape into the cytosol polymerize actin on their surface and use filament assembly to power intracellular actin-based motility (ABM), generating actin comet tails that trail the moving bacteria (reviewed in Haglund and Welch 2011; Welch and Way 2013; Truong, Copeland and Brumell 2014) (Fig. 1). Since the discovery of ABM (Bernardini et al. 1989; Tilney and Portnoy 1989), this phenomenon has captured the attention of researchers in microbiology and cell biology, and studying this process has led to important advances in both fields.
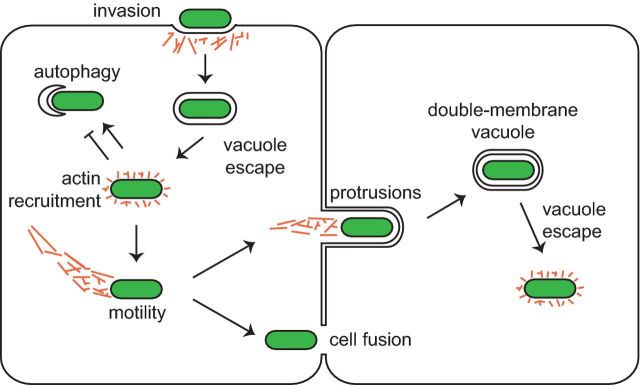
Figure 1.
Actin assembly and ABM play several roles in infection. Actin (orange) is mobilized during multiple stages of infection by intracellular pathogens (green). Actin facilitates bacterial invasion of the host cell. For bacterial pathogens that escape the vacuole into the cytosol, the recruitment of actin and/or actin-interacting proteins on the bacterial surface can influence its susceptibility to autophagy. Moreover, actin assembly powers bacterial ABM through the host cytosol, enabling bacteria to reach the cell periphery, where they enter into protrusions that are engulfed by neighboring cells or promote cell–cell fusion.
Several genera of bacterial pathogens contain species that are capable of ABM (Table 1). These include Shigella and Mycobacteria, for which only one species undergoes ABM, as well as Listeria, Burkholderia and Rickettsia, for which there are two or more species that undergo ABM. Interestingly, different bacterial genera, and even different species within a single genus, have evolved distinct molecular strategies to assemble actin and power motility by mimicking and/or recruiting host actin assembly factors. Understanding these diverse mechanisms can reveal how differences in motility influence pathogenicity.
Table 1.
Bacterial pathogens that undergo ABM.
| Genus | Subgroup | Species | Disease | ABM protein |
|---|---|---|---|---|
| Listeria | n/a | L. monocytogenes | Listeriosis (human) | ActA |
| n/a | L. ivanovii | Listeriosis (ruminant) | ActA | |
| n/a | L. seeligeri | Non-pathogenic | ActA | |
| Burkholderia | n/a | B. pseudomallei | Melioidosis (human) | BimA |
| n/a | B. mallei | Glanders (equine) | BimA | |
| n/a | B. thailandensis | Non-pathogenic | BimA | |
| Rickettsia | SFG | R. rickettsii, R. conorii, R. parkeri, others | Spotted fever and eschar-associated rickettsioses | RickA, Sca2 |
| TRG | R. felis, others | Flea-borne typhus | RickA, Sca2 | |
| TG | R. prowazekii, R. typhi | Epidemic typhus, murine tyhpus | Sca2 (truncated in R. prowazekii) | |
| AG | R. bellii, others | Non-pathogenic | RickA, Sca2 | |
| Shigella | n/a | S. flexneri | Diarrhea in humans | IcsA |
| Mycobacterium | n/a | M. marinum | Skin lesions | Unknown |
In this minireview, we provide an overview of the roles of bacterial ABM in cell–cell spread, autophagy avoidance and virulence. Moreover, we survey host and bacterial actin-polymerizing molecules and the distinct molecular mechanisms by which they harness actin. Finally, we present ABM through a new lens, comparing motility mechanisms between related species of Listeria, Burkholderia and Rickettsia. We intend to open a window into the evolution of ABM mechanisms, the roles of ABM in adapting bacteria to the intracellular environment, and how differences in motility may influence pathogenicity and virulence in humans and other animals.
ROLES OF ABM IN SURVIVAL AND VIRULENCE
ABM influences multiple stages of bacterial infection in the host. One key function of ABM is to promote cell–cell spread (Fig. 1), as mutants that fail to polymerize actin are defective in spread (Bernardini et al. 1989; Domann et al. 1992; Kocks et al. 1992; Kleba et al. 2010; French et al. 2011; Reed et al. 2014). Motility facilitates bacterial movement to the host plasma membrane, where Listeria, Rickettsia and Shigella spp. enter into protrusions in the plasma membrane of the donor cell that can be engulfed by a receiving cell (Tilney and Portnoy 1989; Kadurugamuwa et al. 1991; Gouin et al. 1999; Van Kirk, Hayes and Heinzen 2000; also reviewed in Ireton 2013). In contrast, Burkholderia spp. induce fusion of infected cells with uninfected neighbors to form multinucleated giant cells (MNGCs) (Kespichayawattana et al. 2000; French et al. 2011). The ability of these bacteria to spread directly between host cells or fuse cells without leaving the cell allows them to evade immune defenses that are active in the extracellular environment.
A second and emerging role for pathogen actin assembly and ABM is the manipulation of pathways that target bacteria for destruction via autophagy (reviewed in Mostowy and Shenoy 2015) (Fig. 1). For Listeria monocytogenes, actA mutants are more susceptible to autophagy (Birmingham et al. 2007; Yoshikawa et al. 2009), and ActA is proposed to recruit host actin assembly proteins to protect bacteria from recognition by the autophagy machinery (Yoshikawa et al. 2009). In contrast, Shigella flexneri IcsA stimulates autophagy (Ogawa et al. 2005), and actin assembly by S. flexneri promotes the formation of cages of septin proteins around the bacteria that are important for recruiting the autophagy machinery (Mostowy et al. 2010, 2011). However, S. flexneri also counteracts the autophagy machinery using the type III secreted effector IcsB (Ogawa et al. 2005), which acts together with actin regulators (Baxt and Goldberg 2014) to antagonize the recruitment of autophagy proteins. Thus, the role of actin assembly proteins and ABM in modulating autophagy is complicated and may differ between pathogens. Moreover, whether and how actin recruitment by other pathogens such as Rickettsia, Burkholderia and Mycobacteria affects autophagy remains to be determined.
The roles of ABM in bacterial infection have been revealed by identifying and studying mutations in the genes encoding actin assembly proteins (Table 1). For example, the actA gene in L. monocytogenes is necessary for ABM (Domann et al. 1992; Kocks et al. 1992), and the corresponding gene in L. ivanovii fulfills the same role (Kreft, Dumbsky and Theiss 1995). ActA is also an important virulence factor for L. monocytogenes, as an actA deletion mutation exhibits a 103-fold increased LD50 in mice (Brundage et al. 1993), a 30-fold increased LD50 in zebrafish (Levraud et al. 2009) as well as reduced maternal-fetal transmission in pregnant guinea pigs (Bakardjiev, Stacy and Portnoy 2005) and mice (Le Monnier et al. 2007) (the role of ActA in virulence is also reviewed in Travier and Lecuit 2014). The bimA gene in Burkholderia pseudomallei (Stevens et al. 2005b), B. mallei (Schell et al. 2007) and B. thailandensis (French et al. 2011) is similarly required for actin polymerization. Surprisingly, a B. mallei bimA mutant has an identical LD50 to wild type upon intraperitoneal injection of Syrian hamsters (Schell et al. 2007), and it remains unclear whether bimA is a virulence factor during other routes of infection or for other Burkholderia species. Rickettsia have two genes that are required for different temporal phases of ABM—rickA is required for early-phase ABM of Rickettsia parkeri (Reed et al. 2014) and sca2 is required for late-phase ABM of R. rickettsii (Kleba et al. 2010) and R. parkeri (Reed et al. 2014). Sca2 is required for virulence of R. rickettsii, as a sca2 mutant fails to induce fever in guinea pigs (Kleba et al. 2010), but the role of RickA in virulence remains unclear as there are no published studies using a rickA mutant in an animal model. For S. flexneri, the icsA gene is required and sufficient for actin assembly (Bernardini et al. 1989; Goldberg and Theriot 1995), and necessary for virulence in macaque monkeys (Sansonetti et al. 1991). No genes required for Mycobacterium marinum ABM have yet been identified.
To fully understand the function of ABM proteins in virulence in animal models, we must also take into account alternative roles for these proteins. For example, L. monocytogenes ActA is also required for bacterial aggregation and biofilm formation, and ActA-dependent aggregation enhances bacterial persistence within the mouse intestine and shedding in the feces (Travier et al. 2013). Moreover, R. conorii Sca2 can enable host cell invasion (Cardwell and Martinez 2009), although the role of this activity in animals has not been investigated. Future studies will reveal the mechanistic details of how pathogen ABM proteins harness actin for cell–cell spread and autophagy manipulation, and how their roles of actin mobilization and their alternative roles contribute to virulence and disease.
BACTERIAL MOTILITY FACTORS MIMIC EUKARYOTIC HOST ACTIN NUCLEATORS
The bacterial actin assembly proteins mentioned above—including ActA, BimA, RickA, Sca2 and IcsA—act by recruiting and/or mimicking distinct families of host cell actin-polymerizing proteins (reviewed in Haglund and Welch 2011; Bugalhão, Mota and Franco 2015) (Fig. 2). Some of these host proteins function by accelerating or bypassing the rate-limiting nucleation step of actin assembly, which is the formation of a nucleus consisting of three or more actin monomers (monomeric actin is also called globular or G-actin) (Fig. 2A). Others promote the subsequent elongation of actin filaments (F-actin) at the faster-growing barbed (or plus) ends, and often inhibit the activity of capping proteins that would otherwise terminate the assembly process. During or after assembly, additional proteins further organize F-actin into branched, cross-linked or bundled arrays. Host actin nucleating and elongating factors are divided into several types (reviewed in Campellone and Welch 2010; Firat-Karalar and Welch 2011) that are mentioned below together with their bacterial mimics or binding partners.
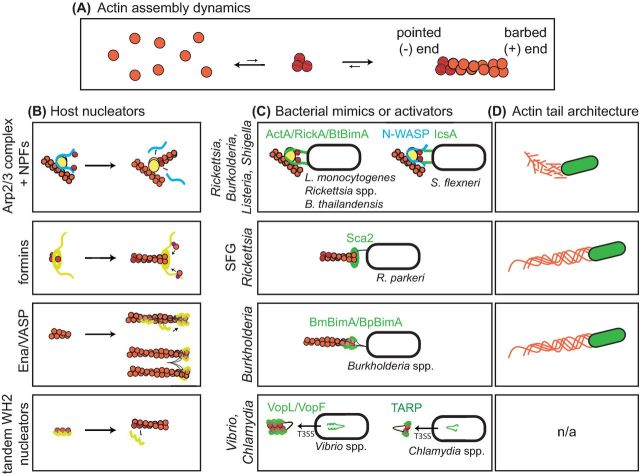
Figure 2.
Eukaryotic actin nucleators and their bacterial mimics. (A) Spontaneous nucleation of actin (orange) involves the formation of a trimer (red), which is kinetically unfavorable. Once a trimer forms, the filament can elongate (or shrink) at both the barbed (+) end or pointed (–) end, although elongation (and shrinking) is faster at the barbed end. (B) Four types of host proteins promote actin nucleation and/or elongation, including the Arp2/3 complex (yellow) and NPFs (blue), formins (yellow) and profilin (magenta), Ena/VASP proteins (yellow), and tandem-WH2-based nucleators (yellow). (C) Bacterial proteins (green) mimic or recruit each different class of host actin-polymerizing proteins (yellow and blue). (D) Representation of actin filament organization in actin tails (orange) corresponding to the bacterial genera and/or species indicated in (C).
One key actin-nucleating factor is the seven-subunit Arp2/3 complex (Fig. 2B). It works together with host proteins called nucleation promoting factors (NPFs) that contain a conserved WCA domain consisting of a G-actin-binding WASP-homology 2 (WH2 or W) sequence(s), and Arp2/3-binding central (C) and acidic (A) sequences. Upon activation by NPFs, the Arp2/3 complex binds to the side of a pre-existing filament and initiates the formation of a new filament that elongates to form a Y-branch (reviewed in Rotty, Wu and Bear 2013). The bacterial actin assembly proteins ActA from Listeria species (Welch et al. 1998; Skoble, Portnoy and Welch 2000; Boujemaa-Paterski et al. 2001; Zalevsky, Grigorova and Mullins 2001), BimA from Burkholderia thailandensis (Sitthidet et al. 2010; Benanti, Nguyen and Welch 2015) and RickA from Rickettsia species (Gouin et al. 2004; Jeng et al. 2004) all contain WCA-like domains that mimic those of host NPFs to activate the Arp2/3 complex (Fig. 2C) (also reviewed in Welch and Way 2013). In contrast, the Shigella flexneri IcsA protein recruits and activates the host NPF N-WASP (Egile et al. 1999). Thus, mimicry or recruitment of NPFs is a common pathogenic strategy to enable ABM.
A second family of host actin-polymerizing proteins is the formins, which promote both nucleation and processive elongation of actin filaments (Fig. 2B). Formins have a conserved formin homology 2 (FH2) domain that forms a homodimeric ring (Xu et al. 2004), which nucleates new actin filaments and then remains associated with the growing barbed ends to accelerate elongation and inhibit capping (reviewed in Paul and Pollard 2009; Chesarone, DuPage and Goode 2010). Formins also have a proline-rich formin homology 1 (FH1) domain that binds to the G-actin-binding protein profilin, and in some cases WH2-like sequences that bind to G-actin, both of which supply a pool of actin to fuel elongation (Paul and Pollard 2009; Chesarone, DuPage and Goode 2010). The bacterial actin assembly protein Sca2 from the spotted fever group (SFG) Rickettsia species Rickettsia parkeri and R. conorii functionally mimics host formins in its ability to enhance nucleation, promote profilin-dependent elongation and inhibit capping (Fig. 2C) (Haglund et al. 2010; Madasu et al. 2013). The Sca2 N-terminal domain is proposed to interact with the C-terminal domain to enable the formin-like activities (Haglund et al. 2010; Madasu et al. 2013), although the N-terminal domain is structurally distinct from the formin FH2 domain (Madasu et al. 2013). Sca2 also has WH2 and proline-rich sequences that participate in filament assembly (Haglund et al. 2010; Madasu et al. 2013). Therefore, although Sca2 from SFG Rickettsia species is a functional mimic of host formins, it is unclear whether it acts by a similar or distinct molecular mechanism. Moreover, both Listeria monocytogenes and S. flexneri use host diaphanous-related formins to facilitate protrusion formation during cell–cell spread (Heindl et al. 2010; Fattouh et al. 2015), suggesting a role for formins in ABM within membrane protrusions at the cell periphery.
A third type of host actin assembly factors is the Ena/VASP family. These multifunctional proteins accelerate barbed-end elongation, antagonize capping proteins and promote filament bundling (Barzik et al. 2005; Hansen and Mullins 2010; Winkelman et al. 2014) (Fig. 2B). Ena/VASP proteins contain a conserved Ena/VASP homology 1 (EVH1) domain that binds to proline-rich sequences in interacting partners to enable localization, a central proline-rich domain that binds to profilin as well as Ena/VASP homology 2 (EVH2) sequences that bind to F-actin and G-actin (reviewed in Bear and Gertler 2009). Their tetrameric structure is also important for the interaction with actin filament ends (Bachmann et al. 1999; Hansen and Mullins 2010). The bacterial actin assembly BimA proteins of the Burkholderia species B. mallei and B. pseudomallei were recently shown to be mimics of Ena/VASP proteins that can promote elongation, antagonize capping protein and bundle F-actin, as well as nucleate actin (Benanti, Nguyen and Welch 2015) (Fig. 2C). These BimA proteins contain one or more WH2 motifs (Stevens et al. 2005a; Sitthidet et al. 2010, 2011) that may be similar to sequences in the EVH2 domain and mediate interactions with G-actin and/or F-actin (Benanti, Nguyen and Welch 2015). Moreover, BimA oligomerization into a trimeric structure is central to actin assembly activity, as is Ena/VASP oligomerization into a tetramer. It remains unclear, however, the precise molecular mechanism by which these BimA proteins mimic the activity of Ena/VASP proteins.
Finally, a fourth type of host actin assembly protein is the tandem-WH2-based nucleators (Fig. 2B). While bacterial mimics of this nucleator family have been identified, none are known to be involved in ABM. However, we include a brief discussion of them in this review to illustrate the full range of diversity observed in bacterial mimics of actin assembly factors. Tandem-WH2-based nucleator proteins have up to four WH2 motifs that bind to G-actin to facilitate nucleation and are also implicated in regulating elongation (reviewed in Carlier et al. 2011; Dominguez 2016). Bacterial mimics of tandem WH2 nucleators include two secreted effector proteins from Vibrio species, VopF from Vibrio cholerae (Tam et al. 2007; Pernier et al. 2013; Avvaru, Pernier and Carlier 2015) and VopL from V. parahaemolyticus (Liverman et al. 2007; Namgoong et al. 2011; Yu et al. 2011; Zahm et al. 2013), as well as TARP from Chlamydia species (Jewett et al. 2006, 2010; Jiwani et al. 2013) (Fig. 2C). Because none of these proteins are implicated in pathogen ABM, they will not be discussed further.
From the examples presented above, it is clear that pathogens have evolved to mimic or recruit all of the major types of host actin nucleation and elongation proteins. Moreover, pathogen mimics of all such host proteins, except the tandem-WH2-based nucleators, have been shown to participate in ABM, and it is possible that tandem-WH2-based mediators of ABM will be discovered. What remains unclear is how mimicry or recruitment of different host actin assembly proteins affects ABM parameters, adapts pathogens to their particular environmental niche and affects virulence in animals.
ABM PROTEINS—ORTHOLOGS, ADAPTATION AND VIRULENCE
Answers to the question of why pathogens have evolved proteins that mimic different host actin assembly factors may come from exploring the unexpected observation that bacterial ABM proteins from related species often exhibit divergent sequences and mechanisms of actin assembly. In this section, we consider sequence and mechanistic differences in orthologs of ABM proteins from selected bacterial genera, and explore how these differences may affect ABM parameters and disease.
Listeria species ActA
The genus Listeria comprises a group of eight Gram-positive soil saprotrophs (Bakker et al. 2010b). Six species are non-pathogenic, including Listeria innocua, L. welshimeri, L. seeligeri, L. marthii, L. rocourtiae and L. grayi. Two species are facultative pathogens—L. monocytogenes is a pathogen of animals including humans, and L. ivanovii is primarily a pathogen of ruminants. Consistent with an important role of ABM in pathogenicity, the pathogenic species L. monocytogenes and L. ivanovii are also the only two observed to undergo ABM (Chakraborty et al. 1995; Gouin et al. 1995; Kreft, Dumbsky and Theiss 1995).
The Listeria actA gene, which is required for ABM, encodes the actin polymerization protein ActA, a transmembrane protein exposed on the bacterial surface. The gene is contained within the prfA virulence gene cluster (Vazquez-Boland et al. 1992), which was suggested by population genetics analysis to be present in the most common recent ancestor of Listeria genus (Bakker et al. 2010a). This cluster was lost in the most recent common ancestors of L. welshimeri and L. marthii, and was lost during the evolution of most L. innocua strains (Bakker et al. 2010a), consistent with the failure to observe ABM for these species. The prfA cluster is retained in L. monocytogenes and L. ivanovii, which undergo ABM, as well as in L. seeligeri, for which ABM has not been observed (Gouin, Mengaud and Cossart 1994) (Fig. 3A). It has been suggested, though, that L. seeligeri exhibits low expression of virulence genes (Gouin, Mengaud and Cossart 1994), hinting that low actA expression may contribute to the failure to observe ABM, and perhaps to a loss of virulence.
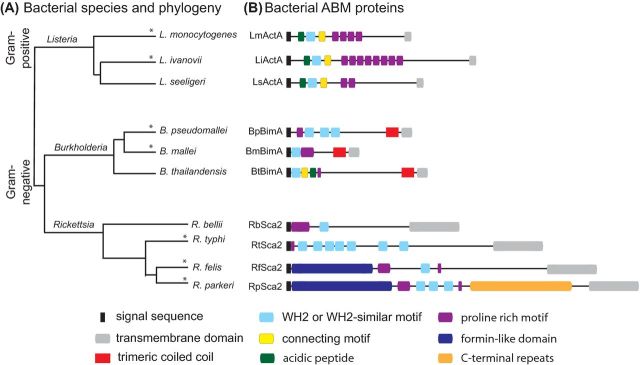
Figure 3.
Species differences in actin assembly proteins. (A) Select species within the genera Listeria, Burkholderia and Rickettsia are represented according to their evolutionary relationships as determined by molecular phylogenetic analyses (branch lengths shown do not indicate evolutionary distances). Asterisk (*) denotes pathogenic species. (B) Schematic representations of bacterial ABM proteins are grouped to show domain comparisons between orthologs (see domain key below).
Interestingly, the amino acid sequence of ActA differs considerably between orthologs from L. monocytogenes (LmActA), L. ivanovii (LiActA) and L. seeligeri (LsActA) (Gouin et al. 1995; Kreft, Dumbsky and Theiss 1995; Gerstel et al. 1996; Müller et al. 2010), with only roughly 20% identity across orthologs. However, all three orthologs retain common sequence features and overall domain organization (Fig. 3B). These include an N-terminal WCA-like domain similar to that of host NPFs, which in LmActA is required for actin nucleation and Y-branch formation with the Arp2/3 complex in vitro (Skoble, Portnoy and Welch 2000; Boujemaa-Paterski et al. 2001) and ABM in cells (Lasa et al. 1997; Pistor et al. 2000; Skoble, Portnoy and Welch 2000). Sequence features also include putative casein kinase 2 (CK2) recognition motifs (Kreft, Dumbsky and Theiss 1995), and phosphorylation by host CK2 is required for efficient ABM and cell–cell spread of L. monocytogenes (Chong et al. 2009). ActA also contains 3–8 central proline-rich motifs (numbers differ between orthologs) that bind to and recruit host Ena/VASP proteins (Chakraborty et al. 1995; Pistor et al. 1995; Gerstel et al. 1996). These are dispensable for ABM but are required for rapid movement (Smith, Theriot and Portnoy 1996; Laurent et al. 1999; Loisel et al. 1999; Auerbuch et al. 2003) and enhanced directional persistence (Auerbuch et al. 2003). The common sequence features in ActA orthologs suggest that they all promote actin nucleation and Y-branching with the Arp2/3 complex and recruit Ena/VASP proteins to promote elongation, thus enabling cooperation between nucleation and elongation factors (reviewed in Chesarone and Goode 2009).
Although all three ActA orthologs likely function by the same molecular mechanism, their significant sequence variability may represent an adaptation of each Listeria species to different hosts or cell types. Because only LmActA has been extensively studied, we do not know how differences in ActA sequence, in particular within the WCA and PR motif regions, influences actin assembly in vitro, ABM in different cell types, or colonization and virulence in different animal models. Elucidating the impact of different orthologs will require replacing the actA gene in one species with orthologs from another species, which has only been done in a single study to show that L. ivanovii actA can restore ABM in an L. monocytogenes actA mutant strain (Gouin et al. 1995). Differences in ActA expression may also contribute to differences in ABM, as was suggested for L. seeligeri (Gouin, Mengaud and Cossart 1994). Thus, it remains to be determined how ActA sequence and expression may adapt different species to their environment or affect virulence and disease.
Burkholderia species BimA
Burkholderia is a genus consisting of more than 40 species of Gram-negative bacteria, most of which are saprophytes. Species in the pseudomallei group are the only ones shown to undergo ABM in host cells (Kespichayawattana et al. 2000; Stevens et al. 2005a). This group includes the human pathogen Burkholderia pseudomallei, which causes melioidosis (Cheng and Currie 2005), and the animal pathogen B. mallei, which causes glanders in equines but can also cause disease in humans (Khan et al. 2013). A third species, B. thailandensis, is not pathogenic to humans or other mammals, but is virulent in Drosophila melanogaster, suggesting that it may be a pathogen of other animal hosts (Pilátová and Dionne 2012). Within the pseudomallei group, B. mallei appears to be a clonal descendant of B. pseudomallei that has undergone genome reduction, whereas B. thailandensis and B. pseudomallei are closely related (Nierman et al. 2004; Kim et al. 2005).
The bimA gene, which encodes the trimeric autotransporter and actin polymerization protein BimA, is retained in all members of the pseudomallei group, but is absent in other Burkholderia species (Fig. 3A). Like other autotransporter proteins, BimA is composed of a C-terminal membrane-anchored autotransporter domain through which the remainder of the protein, or N-terminal passenger domain, is secreted and exposed on the bacterial surface (reviewed in Dautin and Bernstein 2007). The sequence of the BimA passenger domain from B. pseudomallei (BpBimA), B. mallei (BmBimA) and B. thailandensis (BtBimA) is generally highly conserved within each species (Sitthidet et al. 2008), except for BpBimA from a subgroup consisting of 12% of B. pseudomallei strains isolated in Australia, which is nearly identical to BmBimA (Sitthidet et al. 2008). However, BimA differs significantly between species, with only 25%–29% identity between orthologs in the passenger domain. In contrast to Listeria ActA, BimA orthologs have distinct sequence features and overall organization (Fig. 3B). BtBimA contains an N-terminal WCA domain similar to that in host NPFs and activates the host Arp2/3 complex to promote nucleation and Y-branch formation (Stevens et al. 2005a; Sitthidet et al. 2010; Benanti, Nguyen and Welch 2015). In contrast, BpBimA and BmBimA lack the Arp2/3-binding CA sequences and instead contain one (Bm) or three (Bp) WH2 motifs (Stevens et al. 2005a; Benanti, Nguyen and Welch 2015). These WH2 motifs are crucial for BpBimA and BmBimA activities in vitro, which include nucleating actin, binding to barbed ends, promoting elongation, removing capping protein and bundling F-actin, all similar to the activities of host Ena/VASP proteins (Benanti, Nguyen and Welch 2015). Thus, surprisingly, BimA orthologs mimic entirely different classes of host actin-polymerizing proteins, with BtBimA mimicking host NPFs for the Arp2/3 complex, and BpBimA/BmBimA mimicking the Ena/VASP family.
Interestingly, differences in the actin assembly mechanisms of BimA orthologs have been shown to translate into differences in ABM parameters and virulence characteristics. Experiments to discern the functional differences between BimA orthologs began with the successful complementation of the ABM defect of a B. pseudomallei bimA mutant by expression of BtBimA or BmBimA (Stevens et al. 2005a). Later experiments, in which B. thailandensis BimA was replaced with each BimA ortholog, showed that expression of each ortholog causes distinctive ABM parameters and actin filament organization in comet tails (Benanti, Nguyen and Welch 2015) (Fig. 2D). For example, bacteria expressing BtBimA follow more curved trajectories and assemble a dense network of F-actin. In contrast, bacteria expressing BmBimA or BpBimA move in straighter trajectories and assemble bundled F-actin strands. Moreover, bacteria expressing BmBimA are less frequently associated with actin tails, and less efficient at forming plaques in host cell monolayers and inducing MNGC formation. In a hint that BimA ortholog differences may also influence virulence, studies of Australian B. pseudomallei strains that express either BpBimA or BmBimA revealed that there is a significant association between expression of BmBimA and neurological melioidosis, and between expression of BpBimA and pneumonia (Sarovich et al. 2014). Future work using cell culture and animal models, as well as additional epidemiological studies, will reveal whether and how differences in BimA sequence and/or expression influence bacterial adaptation to particular cell types or hosts, and impacts virulence in animals and humans.
Rickettsia species RickA and Sca2
The genus Rickettsia encompasses more than 30 species, all of which are obligate intracellular endosymbionts or parasites. Rickettsia typically lives in arthropod vectors (ticks, fleas, lice and mites). They can be transmitted to mammalian hosts by arthropod bites (reviewed in Azad and Beard 1998; Uchiyama 2012), where they primarily infect endothelial cells, and can also infect macrophages, dendritic cells and other cell types (reviewed in Walker and Ismail 2008). The genus is divided into four groups that differ in host range and in the type and severity of disease caused (if any) (Gillespie et al. 2007, 2008). Members of the SFG, which include Rickettsia rickettsii, R. conorii and R. parkeri, can cause spotted fever disease of varying severity in humans and other mammals (Uchiyama 2012). Within the typhus group (TG), R. prowazekii causes epidemic typhus, whereas R. typhi causes murine typhus (Uchiyama 2012). The transitional group (TRG) species R. felis infects mammals, causing a flea-borne rickettsiosis, and is considered an emerging human pathogen (Brown and Macaluso 2016). Ancestral group (AG) members such as R. bellii are arthropod endosymbionts and are not considered pathogenic. Species within all four groups have been suggested to undergo ABM.
Unlike other bacterial pathogens that undergo ABM, Rickettsia has genes encoding two different actin assembly proteins: RickA and Sca2. The rickA gene is present in SFG, TRG and AG species, but is absent in TG species (McLeod et al. 2004; Walker and Yu 2005). The amino acid sequence of RickA is generally well conserved, with greater than 45% identity between species. All RickA orthologs contain a C-terminal WCA domain similar to that in host NPFs. RickA proteins from the SFG species R. conorii and R. rickettsii were shown to activate the host Arp2/3 complex to promote nucleation and Y-branch formation in vitro (Gouin et al. 2004; Jeng et al. 2004), suggesting the same is true for other orthologs. In the SFG species R. parkeri, RickA was found to mediate an early phase of ABM (occurring 15–60 min post infection) that is characterized by slow and meandering movement and the formation of short and curved actin tails (Reed et al. 2014) (Fig. 2D). One function of early motility appears to be in cell–cell spread, as a rickA::tn mutant forms smaller foci of infection in monolayers of host cells (Reed et al. 2014). Although the importance of RickA in infection of arthropods and mammals remains unclear, the high degree of conservation of RickA sequence in SFG, TRG and AG species suggests that any role in infection is conserved across Rickettsia species. The absence of RickA in TG species is consistent with the fact that they grow to high numbers in host cells and then promote cell lysis and release to enable spread, which may bypass the need for ABM to promote spread by protrusion formation and engulfment.
In contrast with rickA, the sca2 gene, encoding the autotransporter actin assembly protein Sca2, is present in most Rickettsia species. However, sca2 is interrupted in R. peacockii and R. canadensis, and is truncated in R. prowazekii, in keeping with the failure to observe ABM for these species (Heinzen et al. 1993; Baldridge et al. 2005; Simser et al. 2005; Haglund et al. 2010). The amino acid sequence of Sca2 is well conserved within SFG species, with >70% identity throughout the passenger domain, particularly in the N-terminal and C-terminal domains that are necessary for its ability to mimic host formins (Haglund et al. 2010; Cardwell and Martinez 2012; Madasu et al. 2013), and additionally in the central WH2 and proline-rich sequences that participate in actin assembly. Sca2 from TRG species R. felis retains the N-terminal domain of SFG Sca2 (Fig. 3B), but with less sequence conservation, suggesting that it may also be a functional mimic of formins. However, Sca2 orthologs from the TG species R. typhi and the AG species R. bellii are very divergent in that they lack the N-terminal and C-terminal sequences conserved in SFG and TRG species, although they still contain putative WH2 motifs (Fig. 3B) (Haglund et al. 2010; Madasu et al. 2013). This suggests that TG and AG Sca2 orthologs do not mimic host formins, but may instead mimic a different class of host actin-polymerizing proteins, perhaps the tandem WH2 based or Ena/VASP family.
Differences in the actin assembly mechanisms of Sca2 orthologs may translate into differences in ABM parameters. In the SFG species R. parkeri, Sca2 mediates a late phase of ABM (occurring >8–24 h post infection) that is characterized by fast and directionally persistent movement, and the formation of long comet tails consisting of F-actin bundles (Reed et al. 2014) (Fig. 2D). Similar ABM properties and F-actin organization are observed for other SFG species (Heinzen et al. 1993; Gouin et al. 1999; Van Kirk, Hayes and Heinzen 2000), consistent with the high-sequence conservation of SFG Sca2. Although Sca2 from the TRG species R. felis is more divergent, the properties of R. felis ABM have not been described beyond that it appears to form actin tails (Ogata et al. 2005). Interestingly, the AG species R. bellii, which expresses a divergent Sca2, can nevertheless assemble long comet tails of bundled F-actin (Oliver et al. 2014). On the other hand, the TG species R. typhi, which also expresses a divergent Sca2, only infrequently forms actin tails that are shorter than those formed by other species (Teysseire, Chiche-Portiche and Raoult 1992; Heinzen et al. 1993; Van Kirk, Hayes and Heinzen 2000). Thus, although differences in Sca2 appear to be correlated with differences in ABM, more work is needed to better characterize the activity of Sca2 from diverse Rickettsia species to determine the relationship between the molecular mechanisms of actin assembly and specific ABM properties. Unfortunately, such work may be hindered by the relative difficulty of genetic manipulations in Rickettsia species.
Because Sca2-driven ABM is central to virulence for the SFG species R. rickettsii (Kleba et al. 2010), it is reasonable to speculate that differences in ABM characteristics between Rickettsia species will impact virulence. However, the role of ABM in virulence is complicated. The most recent common ancestor of the Rickettsia genus was capable of ABM based on the fact that the AG species R. bellii contains both rickA and sca2 genes and undergoes ABM (Oliver et al. 2014). However, R. bellii is primarily considered a tick endosymbiont and not a pathogen, suggesting that ABM evolved as an adaptation to survival within an arthropod or other host, rather than for virulence humans or other mammals. During the evolution of virulent species, Sca2 both gained and lost functionality. In the TRG and SFG species, Sca2 acquired the N-terminal and C-terminal domains that mimic host formin proteins, and this functionality may be linked to virulence. However, in the TG species R. typhi, Sca2 activity may be reduced because ABM is infrequent (Teysseire, Chiche-Portiche and Raoult 1992; Heinzen et al. 1993; Van Kirk, Hayes and Heinzen 2000), and in R. prowazekii Sca2 is truncated and this species does not undergo ABM. Thus, although ABM mechanisms may play a crucial role in survival and influence virulence for some Rickettsia species, some species have discarded ABM entirely, and in all cases other factors certainly contribute to pathogenicity (Clark et al. 2015).
CONCLUDING REMARKS
The ability to undergo ABM has evolved independently in diverse genera of bacterial pathogens. ABM arose as an adaptation to bacterial life inside the cytosol of diverse eukaryotic host cells, and has been co-opted as an essential virulence strategy during infection of humans and other animals. This process enables cell–cell spread during infection, and may also affect intracellular survival by modulating host autophagy pathways. In recent years, it has become clear that bacterial pathogens have evolved to mimic or usurp the full spectrum of host actin polymerization molecules, including the Arp2/3 complex and NPFs, formins, Ena/VASP proteins and tandem-WH2-based factors. Perhaps more surprisingly, even closely related pathogens can mimic entirely different actin polymerization pathways, suggesting that there is considerable plasticity with regard to ABM mechanisms. The evolution of diversity in ABM proteins sequences and mechanisms of action may be driven by selection for specific ABM characteristics, as well as by selection for alternative roles of ABM proteins in other processes such as invasion or autophagy avoidance. Emerging information suggests that differences in ABM mechanisms may also impact the ability of related species to colonize host organisms, and/or cause disease in humans and animals, perhaps by adapting pathogens to infect different cell types. Future investigations into the diversity of pathogen actin assembly mechanisms, and their contribution to colonization and virulence in different hosts and cell types, will illuminate the evolutionary history of ABM and further our understanding of the selective pressures that influence this important interface between intracellular pathogens and the cytoskeleton of their eukaryotic hosts.
FUNDING
This work was supported by the National Institutes of Health [R01 AI109044, R21 AI109270 and R21 AI119743 to MDW] and the American Heart Association [14PRE18150013 to JEC].
Conflict of interest. None declared.
REFERENCES
- Auerbuch V, Loureiro JJ, Gertler FB, et al. Ena/VASP proteins contribute to Listeria monocytogenes pathogenesis by controlling temporal and spatial persistence of bacterial actin-based motility. Mol Microbiol. 2003;49:1361–75. doi: 10.1046/j.1365-2958.2003.03639.x. [DOI] [PubMed] [Google Scholar]
- Avvaru BS, Pernier J, Carlier MF. Dimeric WH2 repeats of VopF sequester actin monomers into non-nucleating linear string conformations: an X-ray scattering study. J Struct Biol. 2015;190:192–9. doi: 10.1016/j.jsb.2015.03.008. [DOI] [PubMed] [Google Scholar]
- Azad AF, Beard CB. Rickettsial pathogens and their arthropod vectors. Emerg Infect Dis. 1998;4:179–86. doi: 10.3201/eid0402.980205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann C, Fischer L, Walter U, et al. The EVH2 domain of the vasodilator-stimulated phosphoprotein mediates tetramerization, F-actin binding, and actin bundle formation. J Biol Chem. 1999;274:23549–57. doi: 10.1074/jbc.274.33.23549. [DOI] [PubMed] [Google Scholar]
- Bakardjiev AI, Stacy BA, Portnoy DA. Growth of Listeria monocytogenes in the guinea pig placenta and role of cell-to-cell spread in fetal infection. J Infect Dis. 2005;191:1889–97. doi: 10.1086/430090. [DOI] [PubMed] [Google Scholar]
- Bakker den HC, Bundrant BN, Fortes ED, et al. A population genetics-based and phylogenetic approach to understanding the evolution of virulence in the genus Listeria. Appl Environ Microb. 2010a;76:6085–100. doi: 10.1128/AEM.00447-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker den HC, Cummings CA, Ferreira V, et al. Comparative genomics of the bacterial genus Listeria: genome evolution is characterized by limited gene acquisition and limited gene loss. BMC Genomics. 2010b;11:688. doi: 10.1186/1471-2164-11-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldridge GD, Burkhardt N, Herron MJ, et al. Analysis of fluorescent protein expression in transformants of Rickettsia monacensis, an obligate intracellular tick symbiont. Appl Environ Microb. 2005;71:2095–105. doi: 10.1128/AEM.71.4.2095-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M, Kotova TI, Higgs HN, et al. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J Biol Chem. 2005;280:28653–62. doi: 10.1074/jbc.M503957200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxt LA, Goldberg MB. Host and bacterial proteins that repress recruitment of LC3 to Shigella early during infection. PLoS One. 2014;9:e94653. doi: 10.1371/journal.pone.0094653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear JE, Gertler FB. Ena/VASP: towards resolving a pointed controversy at the barbed end. J Cell Sci. 2009;122:1947–53. doi: 10.1242/jcs.038125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benanti EL, Nguyen CM, Welch MD. Virulent Burkholderia species mimic host actin polymerases to drive actin-based motility. Cell. 2015;161:348–60. doi: 10.1016/j.cell.2015.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini ML, Mounier J, d'Hauteville H, et al. Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. P Natl Acad Sci USA. 1989;86:3867–71. doi: 10.1073/pnas.86.10.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birmingham CL, Canadien V, Gouin E, et al. Listeria monocytogenes evades killing by autophagy during colonization of host cells. Autophagy. 2007;3:442–51. doi: 10.4161/auto.4450. [DOI] [PubMed] [Google Scholar]
- Boujemaa-Paterski R, Gouin E, Hansen G, et al. Listeria protein ActA mimics WASp family proteins: it activates filament barbed end branching by Arp2/3 complex. Biochemistry. 2001;40:11390–404. doi: 10.1021/bi010486b. [DOI] [PubMed] [Google Scholar]
- Brown LD, Macaluso KR. Rickettsia felis, an emerging flea-borne rickettsiosis. Curr Trop Med Rep. 2016;3:27–39. doi: 10.1007/s40475-016-0070-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brundage RA, Smith GA, Camilli A, et al. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. P Natl Acad Sci USA. 1993;90:11890–4. doi: 10.1073/pnas.90.24.11890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugalhão JN, Mota LJ, Franco IS. Bacterial nucleators: actin' on actin. Pathog Dis. 2015;73 doi: 10.1093/femspd/ftw078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campellone KG, Welch MD. A nucleator arms race: cellular control of actin assembly. Nat Rev Mol Cell Bio. 2010;11:237–51. doi: 10.1038/nrm2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carabeo R. Bacterial subversion of host actin dynamics at the plasma membrane. Cell Microbiol. 2011;13:1460–9. doi: 10.1111/j.1462-5822.2011.01651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. The Sca2 autotransporter protein from Rickettsia conorii is sufficient to mediate adherence to and invasion of cultured mammalian cells. Infect Immun. 2009;77:5272–80. doi: 10.1128/IAI.00201-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardwell MM, Martinez JJ. Identification and characterization of the mammalian association and actin-nucleating domains in the Rickettsia conorii autotransporter protein, Sca2. Cell Microbiol. 2012;14:1485–95. doi: 10.1111/j.1462-5822.2012.01815.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Husson C, Renault L, et al. Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu. Int Rev Cel Mol Bio. 2011;290:55–85. doi: 10.1016/B978-0-12-386037-8.00005-3. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Ebel F, Domann E, et al. A focal adhesion factor directly linking intracellularly motile Listeria monocytogenes and Listeria ivanovii to the actin-based cytoskeleton of mammalian cells. EMBO J. 1995;14:1314–21. doi: 10.1002/j.1460-2075.1995.tb07117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AC, Currie BJ. Melioidosis: epidemiology, pathophysiology, and management. Clin Microbiol Rev. 2005;18:383–416. doi: 10.1128/CMR.18.2.383-416.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone MA, DuPage AG, Goode BL. Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat Rev Mol Cell Bio. 2010;11:62–74. doi: 10.1038/nrm2816. [DOI] [PubMed] [Google Scholar]
- Chesarone MA, Goode BL. Actin nucleation and elongation factors: mechanisms and interplay. Curr Opin Cell Biol. 2009;21:28–37. doi: 10.1016/j.ceb.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong R, Swiss R, Briones G, et al. Regulatory mimicry in Listeria monocytogenes actin-based motility. Cell Host Microbe. 2009;6:268–78. doi: 10.1016/j.chom.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark TR, Noriea NF, Bublitz DC, et al. Comparative genome sequencing of Rickettsia rickettsii strains that differ in virulence. Infect Immun. 2015;83:1568–76. doi: 10.1128/IAI.03140-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dautin N, Bernstein HD. Protein secretion in gram-negative bacteria via the autotransporter pathway. Annu Rev Microbiol. 2007;61:89–112. doi: 10.1146/annurev.micro.61.080706.093233. [DOI] [PubMed] [Google Scholar]
- Domann E, Wehland J, Rohde M, et al. A novel bacterial virulence gene in Listeria monocytogenes required for host cell microfilament interaction with homology to the proline-rich region of vinculin. EMBO J. 1992;11:1981–90. doi: 10.1002/j.1460-2075.1992.tb05252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R. The WH2 domain and actin nucleation: necessary but insufficient. Trends Biochem Sci. 2016;41:478–90. doi: 10.1016/j.tibs.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egile C, Loisel TP, Laurent V, et al. Activation of the CDC42 effector N-WASP by the Shigella flexneri IcsA protein promotes actin nucleation by Arp2/3 complex and bacterial actin- based motility. J Cell Biol. 1999;146:1319–32. doi: 10.1083/jcb.146.6.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattouh R, Kwon H, Czuczman MA, et al. The diaphanous-related formins promote protrusion formation and cell-to-cell spread of Listeria monocytogenes. J Infect Dis. 2015;211:1185–95. doi: 10.1093/infdis/jiu546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firat-Karalar EN, Welch MD. New mechanisms and functions of actin nucleation. Curr Opin Cell Biol. 2011;23:4–13. doi: 10.1016/j.ceb.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French CT, Toesca IJ, Wu T-H, et al. Dissection of the Burkholderia intracellular life cycle using a photothermal nanoblade. P Natl Acad Sci USA. 2011;108:12095–100. doi: 10.1073/pnas.1107183108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel B, Grobe L, Pistor S, et al. The ActA polypeptides of Listeria ivanovii and Listeria monocytogenes harbor related binding sites for host microfilament proteins. Infect Immun. 1996;64:1929–36. doi: 10.1128/iai.64.6.1929-1936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Beier MS, Rahman MS, et al. Plasmids and rickettsial evolution: insight from Rickettsia felis. PLoS One. 2007;2:e266. doi: 10.1371/journal.pone.0000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JJ, Williams K, Shukla M, et al. Rickettsia phylogenomics: unwinding the intricacies of obligate intracellular life. PLoS One. 2008;3:e2018. doi: 10.1371/journal.pone.0002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MB, Theriot JA. Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. P Natl Acad Sci USA. 1995;92:6572–6. doi: 10.1073/pnas.92.14.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E, Dehoux P, Mengaud J, et al. iactA of Listeria ivanovii, although distantly related to Listeria monocytogenes actA, restores actin tail formation in an L. monocytogenes actA mutant. Infect Immun. 1995;63:2729–37. doi: 10.1128/iai.63.7.2729-2737.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouin E, Egile C, Dehoux P, et al. The RickA protein of Rickettsia conorii activates the Arp2/3 complex. Nature. 2004;427:457–61. doi: 10.1038/nature02318. [DOI] [PubMed] [Google Scholar]
- Gouin E, Gantelet H, Egile C, et al. A comparative study of the actin-based motilities of the pathogenic bacteria Listeria monocytogenes, Shigella flexneri and Rickettsia conorii. J Cell Sci. 1999;112:1697–708. doi: 10.1242/jcs.112.11.1697. [DOI] [PubMed] [Google Scholar]
- Gouin E, Mengaud J, Cossart P. The virulence gene cluster of Listeria monocytogenes is also present in Listeria ivanovii, an animal pathogen, and Listeria seeligeri, a nonpathogenic species. Infect Immun. 1994;62:3550–3. doi: 10.1128/iai.62.8.3550-3553.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Choe JE, Skau CT, et al. Rickettsia Sca2 is a bacterial formin-like mediator of actin-based motility. Nat Cell Biol. 2010;12:1057–63. doi: 10.1038/ncb2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund CM, Welch MD. Pathogens and polymers: microbe-host interactions illuminate the cytoskeleton. J Cell Biol. 2011;195:7–17. doi: 10.1083/jcb.201103148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen SD, Mullins RD. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J Cell Biol. 2010;191:571–84. doi: 10.1083/jcb.201003014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heindl JE, Saran I, Yi C-R, et al. Requirement for formin-induced actin polymerization during spread of Shigella flexneri. Infect Immun. 2010;78:193–203. doi: 10.1128/IAI.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzen RA, Hayes SF, Peacock MG, et al. Directional actin polymerization associated with spotted fever group Rickettsia infection of Vero cells. Infect Immun. 1993;61:1926–35. doi: 10.1128/iai.61.5.1926-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ireton K. Molecular mechanisms of cell-cell spread of intracellular bacterial pathogens. Open Biol. 2013;3:130079–9. doi: 10.1098/rsob.130079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeng RL, Goley ED, D'Alessio JA, et al. A Rickettsia WASP-like protein activates the Arp2/3 complex and mediates actin-based motility. Cell Microbiol. 2004;6:761–9. doi: 10.1111/j.1462-5822.2004.00402.x. [DOI] [PubMed] [Google Scholar]
- Jewett TJ, Fischer ER, Mead DJ, et al. Chlamydial TARP is a bacterial nucleator of actin. P Natl Acad Sci USA. 2006;103:15599–604. doi: 10.1073/pnas.0603044103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jewett TJ, Miller NJ, Dooley CA, et al. The conserved Tarp actin binding domain is important for chlamydial invasion. PLoS Pathog. 2010;6:e1000997. doi: 10.1371/journal.ppat.1000997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiwani S, Alvarado S, Ohr RJ, et al. Chlamydia trachomatis Tarp harbors distinct G and F actin binding domains that bundle actin filaments. J Bacteriol. 2013;195:708–16. doi: 10.1128/JB.01768-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadurugamuwa JL, Rohde M, Wehland J, et al. Intercellular spread of Shigella flexneri through a monolayer mediated by membranous protrusions and associated with reorganization of the cytoskeletal protein vinculin. Infect Immun. 1991;59:3463–71. doi: 10.1128/iai.59.10.3463-3471.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kespichayawattana W, Rattanachetkul S, Wanun T, et al. Burkholderia pseudomallei induces cell fusion and actin-associated membrane protrusion: a possible mechanism for cell-to-cell spreading. Infect Immun. 2000;68:5377–84. doi: 10.1128/iai.68.9.5377-5384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan I, Wieler LH, Melzer F, et al. Glanders in animals: a review on epidemiology, clinical presentation, diagnosis and countermeasures. Transbound Emerg Dis. 2013;60:204–21. doi: 10.1111/j.1865-1682.2012.01342.x. [DOI] [PubMed] [Google Scholar]
- Kim HS, Schell MA, Yu Y, et al. Bacterial genome adaptation to niches: divergence of the potential virulence genes in three Burkholderia species of different survival strategies. BMC Genomics. 2005;6:174. doi: 10.1186/1471-2164-6-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleba B, Clark TR, Lutter EI, et al. Disruption of the Rickettsia rickettsii Sca2 autotransporter inhibits actin-based motility. Infect Immun. 2010;78:2240–7. doi: 10.1128/IAI.00100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kocks C, Gouin E, Tabouret M, et al. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell. 1992;68:521–31. doi: 10.1016/0092-8674(92)90188-i. [DOI] [PubMed] [Google Scholar]
- Kreft J, Dumbsky M, Theiss S. The actin-polymerization protein from Listeria ivanovii is a large repeat protein which shows only limited amino acid sequence homology to ActA from Listeria monocytogenes. FEMS Microbiol Lett. 1995;126:113–21. doi: 10.1111/j.1574-6968.1995.tb07403.x. [DOI] [PubMed] [Google Scholar]
- Lasa I, Gouin E, Goethals M, et al. Identification of two regions in the N-terminal domain of ActA involved in the actin comet tail formation by Listeria monocytogenes. EMBO J. 1997;16:1531–40. doi: 10.1093/emboj/16.7.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent V, Loisel TP, Harbeck B, et al. Role of proteins of the Ena/VASP family in actin-based motility of Listeria monocytogenes. J Cell Biol. 1999;144:1245–58. doi: 10.1083/jcb.144.6.1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Monnier A, Autret N, Join-Lambert OF, et al. ActA is required for crossing of the fetoplacental barrier by Listeria monocytogenes. Infect Immun. 2007;75:950–7. doi: 10.1128/IAI.01570-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levraud J-P, Disson O, Kissa K, et al. Real-time observation of Listeria monocytogenes-phagocyte interactions in living zebrafish larvae. Infect Immun. 2009;77:3651–60. doi: 10.1128/IAI.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liverman AD, Cheng HC, Trosky JE, et al. Arp2/3-independent assembly of actin by Vibrio type III effector VopL. P Natl Acad Sci USA. 2007;104:17117–22. doi: 10.1073/pnas.0703196104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, et al. Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature. 1999;401:613–6. doi: 10.1038/44183. [DOI] [PubMed] [Google Scholar]
- McLeod MP, Qin X, Karpathy SE, et al. Complete genome sequence of Rickettsia typhi and comparison with sequences of other rickettsiae. J Bacteriol. 2004;186:5842–55. doi: 10.1128/JB.186.17.5842-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madasu Y, Suarez C, Kast DJ, et al. Rickettsia Sca2 has evolved formin-like activity through a different molecular mechanism. P Natl Acad Sci USA. 2013;110:E2677–86. doi: 10.1073/pnas.1307235110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Bonazzi M, Hamon MA, et al. Entrapment of intracytosolic bacteria by septin cage-like structures. Cell Host Microbe. 2010;8:433–44. doi: 10.1016/j.chom.2010.10.009. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Sancho-Shimizu V, Hamon MA, et al. p62 and NDP52 proteins target intracytosolic Shigella and Listeria to different autophagy pathways. J Biol Chem. 2011;286:26987–95. doi: 10.1074/jbc.M111.223610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Shenoy AR. The cytoskeleton in cell-autonomous immunity: structural determinants of host defence. Nat Rev Immunol. 2015;15:559–73. doi: 10.1038/nri3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller AA, Schmid MW, Meyer O, et al. Listeria seeligeri isolates from food processing environments form two phylogenetic lineages. Appl Environ Microb. 2010;76:3044–7. doi: 10.1128/AEM.02243-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Namgoong S, Boczkowska M, Glista MJ, et al. Mechanism of actin filament nucleation by Vibrio VopL and implications for tandem W domain nucleation. Nat Struct Mol Biol. 2011;18:1060–7. doi: 10.1038/nsmb.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman WC, DeShazer D, Kim HS, et al. Structural flexibility in the Burkholderia mallei genome. P Natl Acad Sci USA. 2004;101:14246–51. doi: 10.1073/pnas.0403306101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogata H, Renesto P, Audic S, et al. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLoS Biol. 2005;3:e248. doi: 10.1371/journal.pbio.0030248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, et al. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–31. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Oliver JD, Burkhardt NY, Felsheim RF, et al. Motility characteristics are altered for Rickettsia bellii transformed to overexpress a heterologous rickA gene. Appl Environ Microb. 2014;80:1170–6. doi: 10.1128/AEM.03352-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskel. 2009;66:606–17. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernier J, Orban J, Avvaru BS, et al. Dimeric WH2 domains in Vibrio VopF promote actin filament barbed-end uncapping and assisted elongation. Nat Struct Mol Biol. 2013;20:1069–76. doi: 10.1038/nsmb.2639. [DOI] [PubMed] [Google Scholar]
- Pilátová M, Dionne MS. Burkholderia thailandensis is virulent in Drosophila melanogaster. PLoS One. 2012;7:e49745. doi: 10.1371/journal.pone.0049745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pistor S, Chakraborty T, Walter U, et al. The bacterial actin nucleator protein ActA of Listeria monocytogenes contains multiple binding sites for host microfilament proteins. Curr Biol. 1995;5:517–25. doi: 10.1016/s0960-9822(95)00104-7. [DOI] [PubMed] [Google Scholar]
- Pistor S, Grobe L, Sechi AS, et al. Mutations of arginine residues within the 146-KKRRK-150 motif of the ActA protein of Listeria monocytogenes abolish intracellular motility by interfering with the recruitment of the Arp2/3 complex. J Cell Sci. 2000;113:3277–87. doi: 10.1242/jcs.113.18.3277. [DOI] [PubMed] [Google Scholar]
- Reed SCO, Lamason RL, Risca VI, et al. Rickettsia actin-based motility occurs in distinct phases mediated by different actin nucleators. Curr Biol. 2014;24:98–103. doi: 10.1016/j.cub.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotty JD, Wu C, Bear JE. New insights into the regulation and cellular functions of the ARP2/3 complex. Nat Rev Mol Cell Bio. 2013;14:7–12. doi: 10.1038/nrm3492. [DOI] [PubMed] [Google Scholar]
- Sansonetti PJ, Arondel J, Fontaine A, et al. OmpB (osmo-regulation) and icsA (cell-to-cell spread) mutants of Shigella flexneri: vaccine candidates and probes to study the pathogenesis of shigellosis. Vaccine. 1991;9:416–22. doi: 10.1016/0264-410x(91)90128-s. [DOI] [PubMed] [Google Scholar]
- Sarovich DS, Price EP, Webb JR, et al. Variable virulence factors in Burkholderia pseudomallei (melioidosis) associated with human disease. PLoS One. 2014;9:e91682. doi: 10.1371/journal.pone.0091682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schell MA, Ulrich RL, Ribot WJ, et al. Type VI secretion is a major virulence determinant in Burkholderia mallei. Mol Microbiol. 2007;64:1466–85. doi: 10.1111/j.1365-2958.2007.05734.x. [DOI] [PubMed] [Google Scholar]
- Simser JA, Rahman MS, Dreher-Lesnick SM, et al. A novel and naturally occurring transposon, ISRpe1 in the Rickettsia peacockii genome disrupting the rickA gene involved in actin-based motility. Mol Microbiol. 2005;58:71–9. doi: 10.1111/j.1365-2958.2005.04806.x. [DOI] [PubMed] [Google Scholar]
- Sitthidet C, Korbsrisate S, Layton AN, et al. Identification of motifs of Burkholderia pseudomallei BimA required for intracellular motility, actin binding, and actin polymerization. J Bacteriol. 2011;193:1901–10. doi: 10.1128/JB.01455-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C, Stevens JM, Chantratita N, et al. Prevalence and sequence diversity of a factor required for actin-based motility in natural populations of Burkholderia species. J Clin Microbiol. 2008;46:2418–22. doi: 10.1128/JCM.00368-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sitthidet C, Stevens JM, Field TR, et al. Actin-based motility of Burkholderia thailandensis requires a central acidic domain of BimA that recruits and activates the cellular Arp2/3 complex. J Bacteriol. 2010;192:5249–52. doi: 10.1128/JB.00608-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoble J, Portnoy DA, Welch MD. Three regions within ActA promote Arp2/3 complex-mediated actin nucleation and Listeria monocytogenes motility. J Cell Biol. 2000;150:527–38. doi: 10.1083/jcb.150.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GA, Theriot JA, Portnoy DA. The tandem repeat domain in the Listeria monocytogenes ActA protein controls the rate of actin based motility, the percentage of moving bacteria, and the localization of vasodilator-stimulated phosphoprotein and profilin. J Cell Biol. 1996;135:647–60. doi: 10.1083/jcb.135.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens JM, Ulrich RL, Taylor LA, et al. Actin-binding proteins from Burkholderia mallei and Burkholderia thailandensis can functionally compensate for the actin-based motility defect of a Burkholderia pseudomallei bimA mutant. J Bacteriol. 2005a;187:7857–62. doi: 10.1128/JB.187.22.7857-7862.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens MP, Stevens JM, Jeng RL, et al. Identification of a bacterial factor required for actin-based motility of Burkholderia pseudomallei. Mol Microbiol. 2005b;56:40–53. doi: 10.1111/j.1365-2958.2004.04528.x. [DOI] [PubMed] [Google Scholar]
- Tam VC, Serruto D, Dziejman M, et al. A type III secretion system in Vibrio cholerae translocates a formin/spire hybrid-like actin nucleator to promote intestinal colonization. Cell Host Microbe. 2007;1:95–107. doi: 10.1016/j.chom.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Teysseire N, Chiche-Portiche C, Raoult D. Intracellular movements of Rickettsia conorii and R. typhi based on actin polymerization. Res Microbiol. 1992;143:821–9. doi: 10.1016/0923-2508(92)90069-z. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA. Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol. 1989;109:1597–608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travier L, Guadagnini S, Gouin E, et al. ActA promotes Listeria monocytogenes aggregation, intestinal colonization and carriage. PLoS Pathog. 2013;9:e1003131. doi: 10.1371/journal.ppat.1003131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travier L, Lecuit M. Listeria monocytogenes ActA: a new function for a “classic” virulence factor. Curr Opin Microbiol. 2014;17:53–60. doi: 10.1016/j.mib.2013.11.007. [DOI] [PubMed] [Google Scholar]
- Truong D, Copeland JW, Brumell JH. Bacterial subversion of host cytoskeletal machinery: hijacking formins and the Arp2/3 complex. Bioessays. 2014;36:687–96. doi: 10.1002/bies.201400038. [DOI] [PubMed] [Google Scholar]
- Uchiyama T. Tropism and pathogenicity of rickettsiae. Front Microbiol. 2012;3:230. doi: 10.3389/fmicb.2012.00230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Kirk LS, Hayes SF, Heinzen RA. Ultrastructure of Rickettsia rickettsii actin tails and localization of cytoskeletal proteins. Infect Immun. 2000;68:4706–13. doi: 10.1128/iai.68.8.4706-4713.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kocks C, Dramsi S, et al. Nucleotide sequence of the lecithinase operon of Listeria monocytogenes and possible role of lecithinase in cell-to-cell spread. Infect Immun. 1992;60:219–30. doi: 10.1128/iai.60.1.219-230.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker DH, Ismail N. Emerging and re-emerging rickettsioses: endothelial cell infection and early disease events. Nat Rev Microbiol. 2008;6:375–86. doi: 10.1038/nrmicro1866. [DOI] [PubMed] [Google Scholar]
- Walker DH, Yu X-J. Progress in rickettsial genome analysis from pioneering of Rickettsia prowazekii to the recent Rickettsia typhi. Ann NY Acad Sci. 2005;1063:13–25. doi: 10.1196/annals.1355.003. [DOI] [PubMed] [Google Scholar]
- Welch MD, Rosenblatt J, Skoble J, et al. Interaction of human Arp2/3 complex and the Listeria monocytogenes ActA protein in actin filament nucleation. Science. 1998;281:105–8. doi: 10.1126/science.281.5373.105. [DOI] [PubMed] [Google Scholar]
- Welch MD, Way M. Arp2/3-mediated actin-based motility: a tail of pathogen abuse. Cell Host Microbe. 2013;14:242–55. doi: 10.1016/j.chom.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman JD, Bilancia CG, Peifer M, et al. Ena/VASP Enabled is a highly processive actin polymerase tailored to self-assemble parallel-bundled F-actin networks with fascin. P Natl Acad Sci USA. 2014;111:4121–6. doi: 10.1073/pnas.1322093111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Moseley JB, Sagot I, et al. Crystal structures of a formin homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116:711–23. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y, Ogawa M, Hain T, et al. Listeria monocytogenes ActA-mediated escape from autophagic recognition. Nat Cell Biol. 2009;11:1233–40. doi: 10.1038/ncb1967. [DOI] [PubMed] [Google Scholar]
- Yu B, Cheng H-C, Brautigam CA, et al. Mechanism of actin filament nucleation by the bacterial effector VopL. Nat Struct Mol Biol. 2011;18:1068–74. doi: 10.1038/nsmb.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahm JA, Padrick SB, Chen Z, et al. The bacterial effector VopL organizes actin into filament-like structures. Cell. 2013;155:423–34. doi: 10.1016/j.cell.2013.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J, Grigorova I, Mullins RD. Activation of the Arp2/3 complex by the Listeria ActA protein. ActA binds two actin monomers and three subunits of the Arp2/3 complex. J Biol Chem. 2001;276:3468–75. doi: 10.1074/jbc.M006407200. [DOI] [PubMed] [Google Scholar]