FIGURE 1.
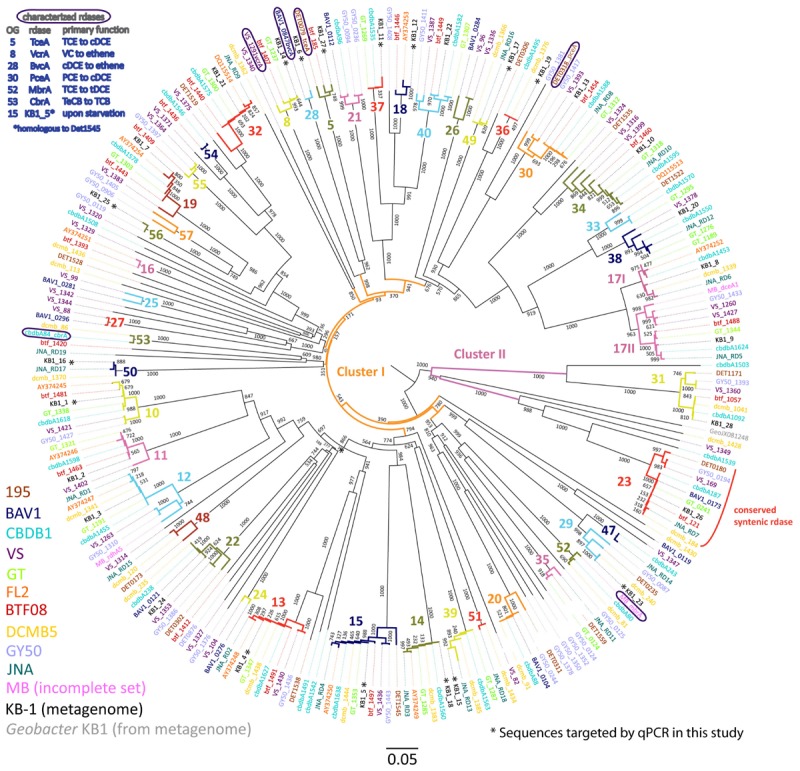
Phylogenetic tree of Dehalococcoides mccartyi reductive dehalogenases based on amino acid sequences. Tree includes sequences predicted from 248 rdhA genes found in eleven isolated D. mccartyi strains as well as those found in the mixed dechlorinating culture KB-1, including the rdhA gene sequence found in Geobacter KB-1 as an outgroup. The scale is the number of substitutions per amino acid. Bootstraps are shown at tree nodes. Sequences names are colored corresponding to their source as shown in the legend at bottom left. The numbers and different branch colors highlight ortholog groups (OGs) where sequences have >90% PID. Sequences corresponding to characterized RDases are circled and their substrates shown in the top right legend (chlorinated substrate abbreviations as in text, plus TeCB, tetrachlorobenzene; TCB, trichlorobenzene).
