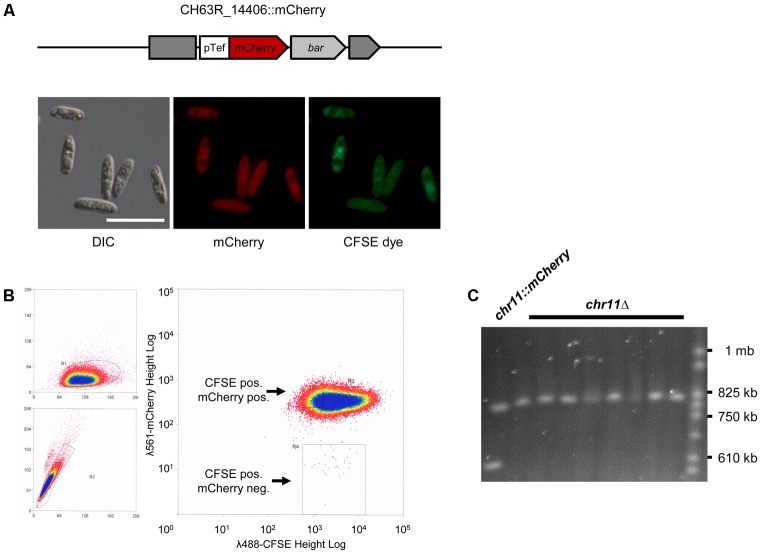
FIGURE 2.
A chromosome loss assay using a targeted insertion of an mCherry gene. (A) Schematic representation of the CH63R_14406 locus on chromosome 11 after insertion of an mCherry reporter cassette by homologous recombination (strain CY7466). The mCherry gene was driven by a strong promotor (pTef). The bar gene conferring bialaphos resistance was used as a selectable marker. The dark gray blocks represent the disrupted CH63R_14406 locus. The lower images show the signals of mCherry in the conidial cytoplasm and of the fluorescent cell tracer CFSE in conidia of this strain. DIC, differential interference contrast, scale bar = 20 μm. (B) Flow cytometry of chr11::mCherry strain (CY7466) used to sort for mCherry negative conidia. We found a loss rate of 3 × 10-4 for chromosome 11. Left: forward and side scatter of cells. FSC-Height/ SSC-Height (top) and SSC-Height/SSC-Area (bottom). Right: fluorescent signals of CFSE at λ = 488 nm (x-axis) and mCherry at λ = 561 (y-axis). Scales are logarithmic and were adjusted to fit the range of fluorescence in the sample. The region gated for isolating mCherry neg. cells is shown as a rectangle. (C) Pulsed-field gel electrophoresis of chromosomes from parental chr11::mCherry conidia and from randomly chosen single colonies of sorted mCherry negative cells. Yeast chromosomes were used as a size marker (CHEF DNA Size Marker).