Figure 5.
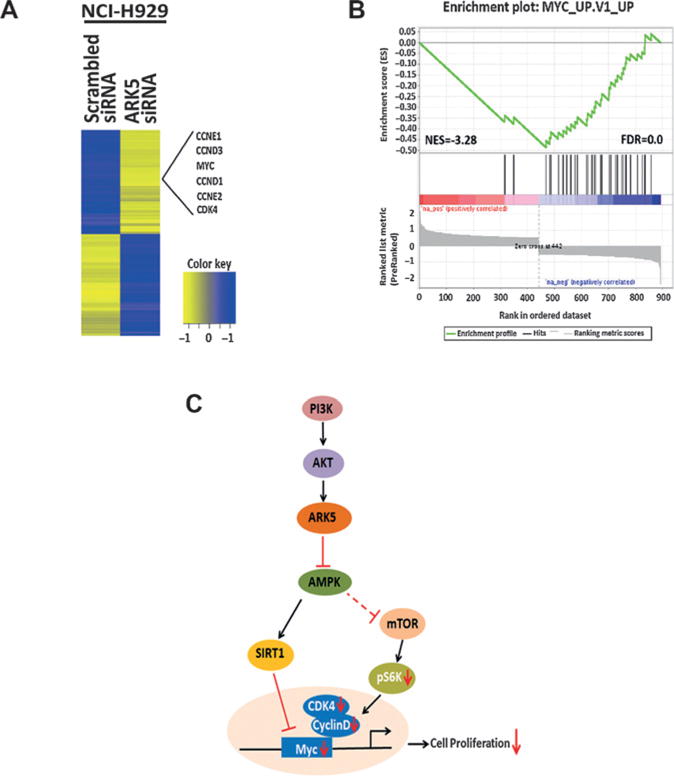
Transcriptomic profiling reveals ARK5 depletion results in the suppression of cell cycle regulating genes in multiple myeloma cell lines. A, RNA sequencing data from ARK5-depleted NCI-H929 cells identified 894 DEGs between ARK5 siRNA and scrambled siRNA. Cell-cycle pathway was enriched in ARK5-depleted cells with cell-cycle regulators like MYC, cyclin D and E, and CDK4 genes significantly repressed after ARK5 depletion as shown. B, enrichment score plot by GSEA showed a strong negative enrichment of MYC target genes in ARK5 depleted NCI-H929 cells. Preranked list of 894 DEGs was used for GSEA in the C6 oncogenic signatures. The distribution of MYC target genes (black lines) is presented as a function of the change in expression between ARK5 siRNA and scrambled siRNA, from highly upregulated (red) to highly downregulated (blue). Normalized enrichment score (NES) and FDR q values for MYC target gene set are shown. C, schematic representing the downstream signaling events of ARK5 is shown.
