Figure 3.
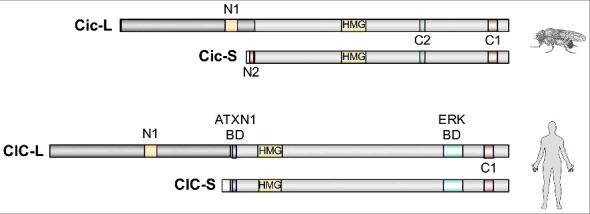
Structure and conservation of CIC orthologs from Drosophila and humans. Both species express CIC-L and CIC-S isoforms with alternative N-terminal regions. These isoforms display overlapping distributions in multiple tissues (particularly in mammals [22,25]), although very little is known about the mechanisms controlling these patterns of expression both in Drosophila and in mammals. Studies in Drosophila have revealed a key difference between Cic-S and Cic-L: Cic-S contains a specific motif called N2 which is critical for Gro-mediated repression in the embryo [24]. Conversely, the Cic-L isoforms share a domain of unknown function (N1) in their N-terminal regions [23,24]. Other than that, the functional differences between these isoforms remain poorly understood. All short and long isoforms share the HMG-box and C1 domains involved in DNA binding. Although the ATXN1 binding domain (BD) characterized in mammalian CIC proteins [23,26] is only moderately conserved in Drosophila Cic [23] (not shown), Ataxin-1 has been identified in a screen for Cic interactors in Drosophila embryos [18]. The C2 MAPK docking site of Drosophila Cic and a distinct ERK BD of human CIC are also indicated [5,27].
