Figure 2. SG-APEX Identifies Known and Previously Unknown SG Proteins within a Dense Interaction Network.
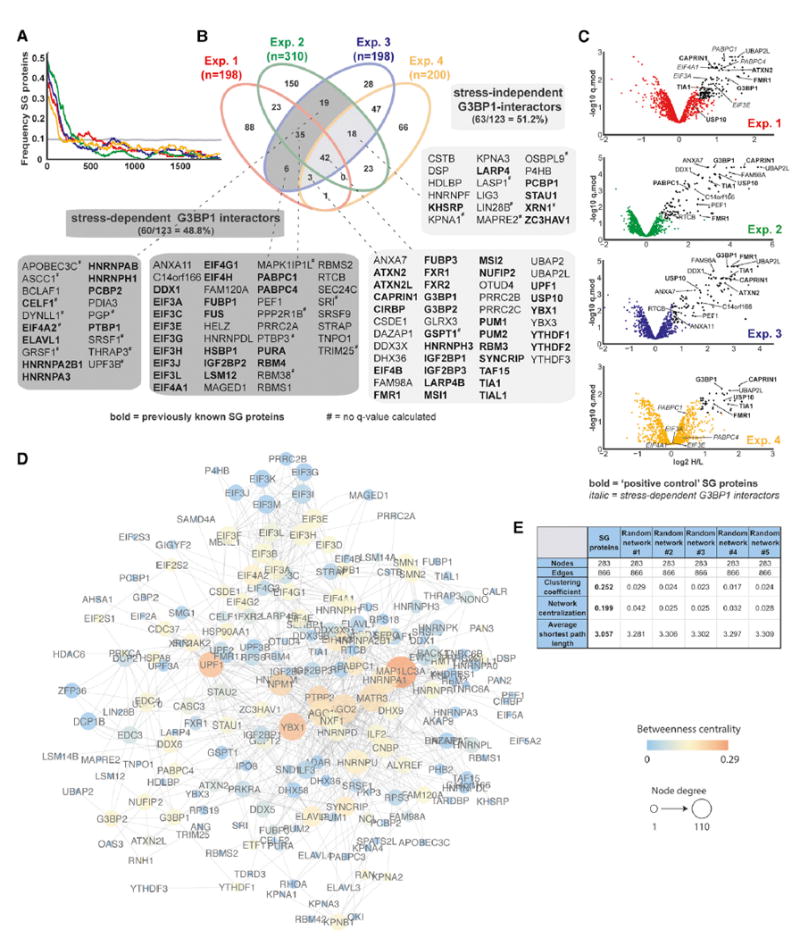
(A) Enrichment frequency distribution of known SG proteins in log2 H/L-ranked proteomics datasets. The dashed line represents 2 times the background frequency of SG proteins across all detected proteins.
(B) Venn diagram showing overlapping hits from four experimental designs, with previously known SG proteins highlighted in bold.
(C) Volcano plots showing statistically significant enrichment of selected known and previously unknown SG proteins across experiments.
(D) Protein interaction network (PIN) of 283 proteins identified as APEX hits in HEK293T cells or previously shown to associate with SGs. Network was visualized in Cytoscape using a force-directed layout.
(E) Common network parameters for the SG-PIN compared to five PINs from a randomly selected equal number of nodes and edges. See also Table S3.
