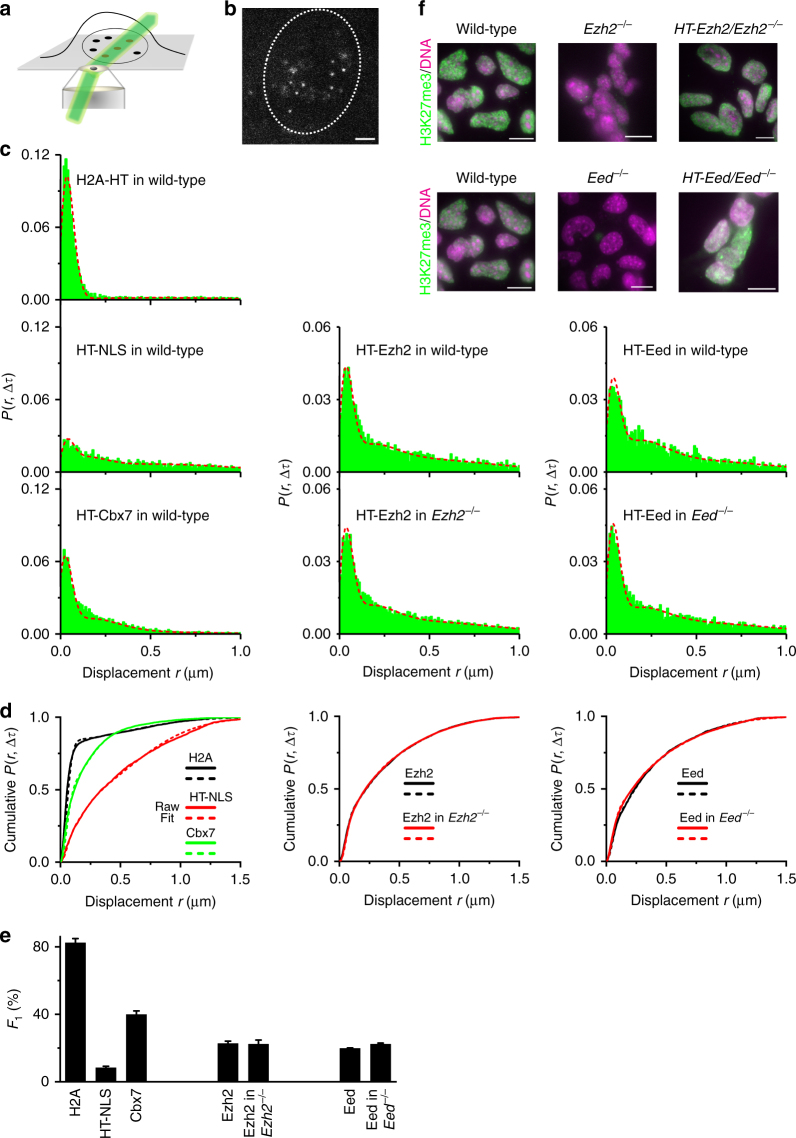
Fig. 1.
PRC2 and Cbx7 exhibit distinct capacities for binding to chromatin. a Schematic illustrating HILO (highly inclined and laminated optical sheet). b Example image showing single HaloTag-Ezh2 molecules labeled with JF549 dye during a 30 ms exposure time. The nucleus was marked by oval white dash circle. The individual white points represent single HaloTag-Ezh2 molecules. Scale bar, 2.0 µm. c Displacement histograms for H2A-HaloTag (N = 52 cells, n = 6172 displacements), HaloTag-NLS (N = 66 cells, n = 6587 displacements), and HaloTag-Cbx7 (N = 40 cells, n = 7725 displacements) in wild-type mES cells, for HaloTag-Ezh2 in wild-type (N = 30 cells, n = 5532 displacements) and Ezh2−/− (N = 50 cells, n = 14,427 displacements) mES cells, and for HaloTag-Eed in wild-type (N = 36 cells, n = 5444 displacements) and Eed−/− (N = 77 cells, n = 8845 displacements) mES cells. The short dash red curve indicates the overall fit. NLS, nuclear localization sequence. HT, HaloTag. d Cumulative distribution of displacements for H2A-HaloTag, HaloTag-NLS, and HaloTag-Cbx7 in wild-type mES cells, for HaloTag-Ezh2 in wild-type and Ezh2−/− mES cells, and for HaloTag-Eed in wild-type and Eed−/− mES cells. The cumulative distributions were fitted with two or three components. Fitted parameters are shown in Supplementary Table 1. Unless otherwise indicated, the reported kinetic fractions and diffusion constants were obtained from the cumulative distributions. Solid curve represents raw data. Short dash curve is fitted data. e Fraction of the chromatin-bound population (F1) obtained from d. Results are means ± SD from three biological replicates. f Immunostaining of H3K27me3 in wild-type, Ezh2─/─, Eed─/─, HaloTag-Ezh2/Ezh2─/─, and HaloTag-Eed/Eed─/─ mES cells by using antibody directed against H3K27me3 (green). DNA was stained with hoechst (red). Overlay images are shown. The residual H3K27me3 level was detectable in Ezh2─/─ mES cells because of the presence of Ezh1. Scale bar, 5.0 µm