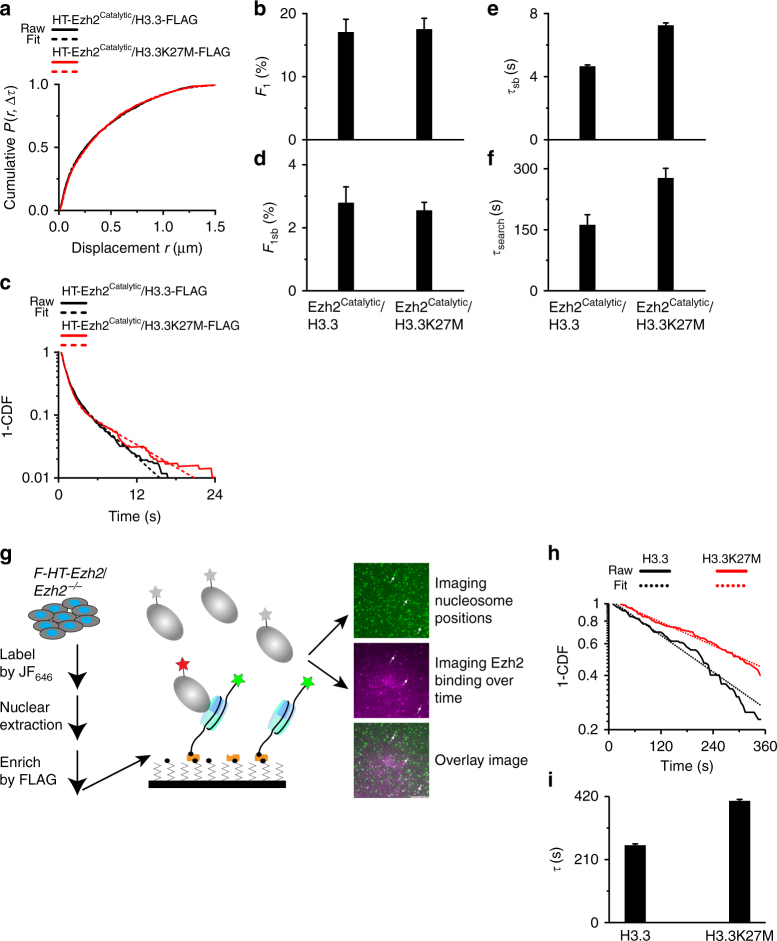
Fig. 6.
The interactions between H3.3K27M and Ezh2 stabilize Ezh2 on chromatin. a Cumulative distribution of displacements for HaloTag-Ezh2Catalytic in HEK293T cells expressing H3.3-FLAG (N = 42 cells, n = 2365 displacements) or H3.3K27M-FLAG (N = 36 cells, n = 4120 displacements). The cumulative distributions were fitted with three populations. Ezh2Catalytic contains only the catalytic lobe (Fig. 3a). Fitted parameters are shown in Supplementary Table 7. b Fraction of the chromatin-bound population (F1) obtained from a. Results are means ± SD. c Survival probability distribution of the dwell times for HaloTag-Ezh2Catalytic in HEK293T cells expressing H3.3-FLAG (N = 56 cells, n = 1412 displacements) or H3.3K27M-FLAG (N = 46 cells, n = 1042 displacements). The distributions were fitted with a two-component exponential decay model. Fitted parameters are shown in Supplementary Table 8. d–f Fraction (d), residence time (e), and search time (f) of the long-lived population (F1sb) for HaloTag-Ezh2Catalytic in HEK293T cells expressing H3.3-FLAG or H3.3K27M-FLAG. Results are means ± SD. g In vitro single-molecule binding assay. FLAG-HaloTag-Ezh2 (F-HT-Ezh2) was expressed stably in Ezh2─/─ mES cells and labeled by JF646 dyes. FLAG-HaloTag-Ezh2-PRC2 was one-step enriched by FLAG antibody and loaded into the flow cells. Nucleosome labeled with Alexa Fluor®488 was tethered to the surface of a coverslip that had been passivated and functionalized with NeutrAvidin. The position of individual nucleosomes was recorded (green image). We followed the duration of FLAG-HaloTag-Ezh2-PRC2 on nucleosome (magenta image). The colocalization of nucleosome and FLAG-HaloTag-Ezh2 indicates a binding event (overlay image). Arrowheads indicate binding events. Scale bar, 5.0 µm. h Survival probability distribution of the dwell times for FLAG-HaloTag-Ezh2-PRC2 on H3.3-nucleosome (n = 99 trajectories) and on H3.3K27M-nucleosome (n = 115 trajectories). The distributions were fitted with a one-component exponential decay model. i Residence time of FLAG-HaloTag-Ezh2-PRC2 on H3.3-nucleosome and on H3.3K27M-nucleosome. Results are means ± SD from three biological replicates