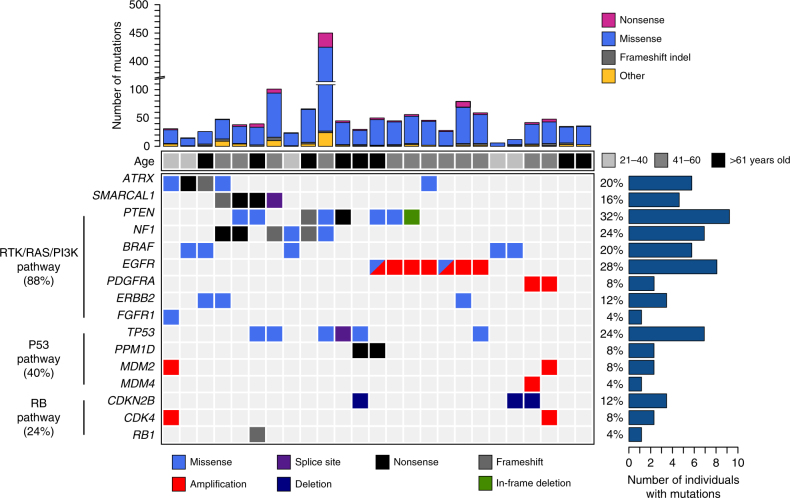
Fig. 1.
The mutational landscape of somatic coding alterations in TERTpWT-IDHWT GBM. Whole exome sequencing was performed on TERTpWT-IDHWT GBMs (N = 25). Recurrently mutated pathways identified included the RTK/RAS/PI3K (88%), P53 (40%), and RB (24%) pathways. Somatic mutation rates per case are shown with corresponding patient age (top). Recurrently mutated genes displayed determined to be significantly mutated (IntOgen algorithm, P < 0.05, n ≥ 2) are shown, as well as select lower frequency genes that are recurrently mutated in glioma or known oncogenes/tumor suppressors in the pathways shown. The mutation frequency of each gene is shown (right) as a percentage of the total cohort