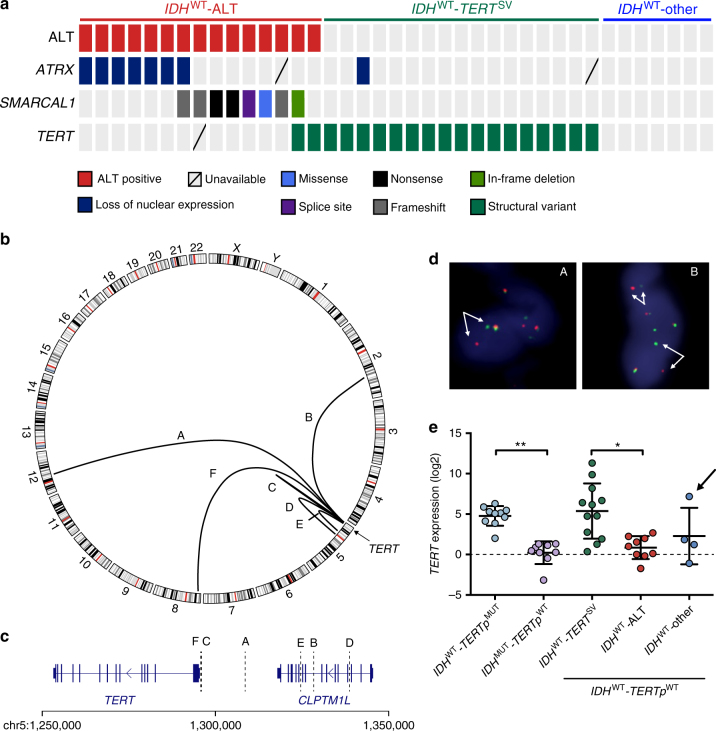
Fig. 2.
Inactivating mutations in SMARCAL1 and ATRX, and rearrangements upstream of TERT are frequent in TERTpWT-IDHWT GBMs and related to distinct telomere maintenance mechanisms. a Based on ALT assessment by both telomere FISH and C-circle (dot blot), 38.5% (15/39) of TERTpWT-IDHWT GBMs exhibit signs of ALT. Of these, approximately half exhibit loss of ATRX expression (IHC) and half harbor mutations in SMARCAL1, in a largely mutually exclusive manner. TERT rearrangements were identified by whole genome sequencing (N = 8). Break-apart FISH was used to screen the cohort for TERT rearrangements, which were present in 50% (19/38) of all TERTpWT-IDHWT GBMs. b Circos plot of rearrangements identified upstream of TERT by whole genome sequencing of ALT-negative GBMs (N = 8). Several cases were interchromosomal translocations (A, B, F), while the remaining cases were intrachromosomal (C, D, E). c The breakpoints of the rearrangements identified by whole genome sequencing span a region in the 50 kb upstream of TERT. d Examples of FISH on patient tumor tissue showing break-apart signal, indicating TERT-rearrangement. Arrows identify break-apart signals. e TERT expression was assessed by rt-qPCR relative to GAPDH. IDHWT-TERTSV (n = 12) tumors exhibit significantly higher TERT expression than the IDHWT-ALT subgroup (n = 9, P < 0.05). This is a similar trend seen among known GBM groups, where the IDHWT-TERTpMUT GBMs (telomerase positive) exhibit increased TERT expression compared to IDHMUT-TERTpWT (ALT positive) GBMs (P < 0.01). The IDHWT-other subgroup is ALT negative, but does not harbor detectable TERT rearrangements. One case in this group harbors MYC amplification (arrow), known to increase TERT expression due to the presence of MYC binding sites in the TERT promoter region. Error bars in e denote s.d. *P < 0.05; **P < 0.01; Kruskal–Wallis test with Dunn’s multiple comparisons test. Three technical replicates were used for TERT mRNA expression