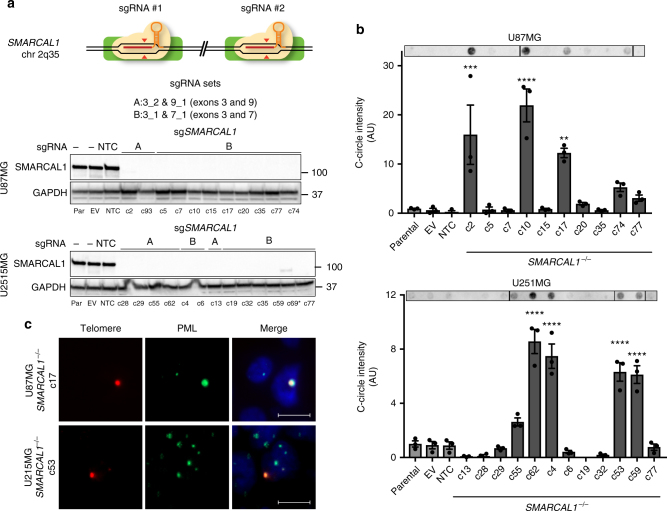
Fig. 5.
Loss of SMARCAL1 in glioblastoma cell lines leads to features of ALT. a CRISPR/Cas9 gene editing was used to generate SMARCAL1 knockout GBM lines (U87MG and U251MG). Two guide combinations (A: 3_2 & 9_1 and B: 3_1 & 7_1) were used targeting exons 3 and 9 and 3 and 7, respectively. Clones were sequenced and validated as isogenic knockout lines by western blot (*clone c69 was excluded due to faint band). b Cell lines were assessed for C-circle accumulation (by dot blot), a characteristic observed in cells using ALT for telomere maintenance. Approximately 30% of isogenic SMARCAL1 knockout GBM lines isolated in both U87MG and U251MG exhibited significantly increased levels of C-circles (U87MG: 4/12, U251MG: 3/10), as compared to the parental cell line. c C-circle-positive SMARCAL1 knockout clones were assessed for the presence of ALT-associated PML bodies (APBs), as indicated by the co-localization of PML (immunofluorescence) and ultrabright telomere foci (FISH). Rare cells were identified in these C-circle-positive clones with APBs. Error bars in b denote s.e.m. **P < 0.01; ***P < 0.001; ****P < 0.0001; one-way ANOVA with Dunnett’s multiple comparisons test relative to parental cell line. Scale bar indicates 10 μm. C-circle experiments were performed in triplicate