Table 2.
Haplotype analysis in cases with the c.1073dup mutation in ANTXR2 gene
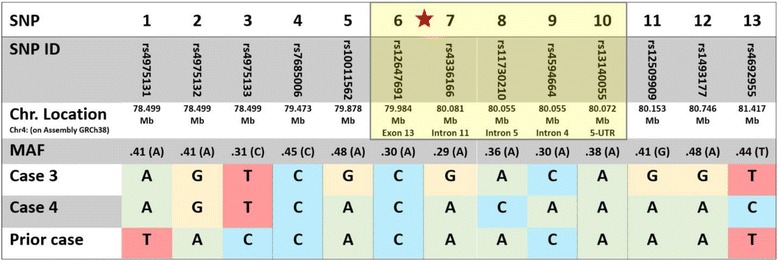
Haplotype analysis was performed in three cases of HFS homozygous for the c.1073dup (p.Ala359Cysfs*13) mutation in ANTXR2, including Case 3 and Case 4 from the present report, and a previously reported patient of Iranian descent with the identical mutation. Thirteen informative single nucleotide polymorphisms (SNPs) with minor allele frequencies (MAF) ranging from 0.29 to 0.48 in a 2.91 Mb region of chromosome 4 (https://www.ncbi.nlm.nih.gov/SNP) were sequenced in each case, including five SNPs within the ANTXR2 gene (nos. 6-10, yellow highlight). The position of the c.1073dup mutation is indicated by a red asterisk. Analysis revealed that while each patient was homozygous for the allele of each included SNP as expected in consanguineous families, the SNP haplotype differed between these three cases. These data suggest an independent origin of this mutation event rather than being inherited from a common ancestor
