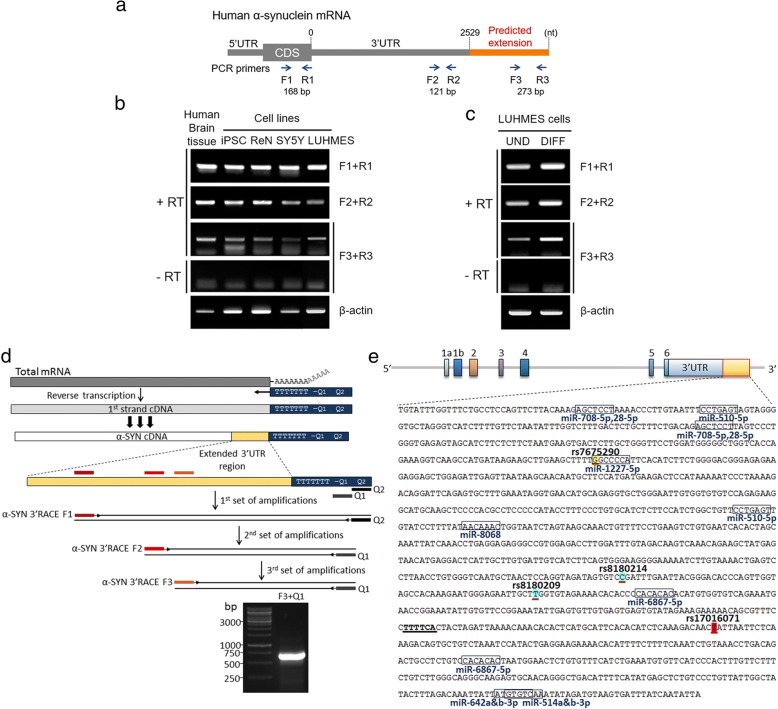
Fig. 2.
PCR amplification and the sequence of the extended 3′UTR of α-SYN mRNA and the potential regulatory miRNAs and SNPs. a The schematic structure of α-SYN mRNA including the predicted extension of 3′UTR. Three different primers sets for PCR amplification are shown. CDS; coding DNA sequence. b RT-PCR for the extended 3′UTR in the SN tissue of postmortem brains (one representative sample is shown here out of 8 brain samples) and other human neuronal cell lines: iPSC, dopaminergic neurons (DIV 60) differentiated from induced pluripotent stem cells; ReN, human ventral mesencephalic neuronal progenitor cells; SY5Y, human neuroblastoma cells; LUHMES, immortalized human dopaminergic neuronal precursor cells. “+” or “-” RT; with or without RT reaction. c Expression of the extended 3′UTR in undifferentiated (UND) and differentiated (DIFF) LUHMES cells. d Schematic overview of the 3′-RACE procedure. Three serial amplification steps using three forward and two reverse primers were performed to amplify the terminal region of extended α-SYN 3′UTR. e Schematics of SNCA gene structure including the newly identified end of the last exon with yellow box. The sequence of the extended 3′UTR with marks for binding sites of miRNAs and SNPs are shown. miRNAs with the high target prediction scores (> 70) are marked. The four SNPs highlighted in the extended 3′UTR, are in significant linkage disequilibrium (r2 ≥ 0.95) with the PD-implicated SNP (rs11931074) in various populations. The distances of these indicated SNPs from the lead SNP (rs11931074) are as follows: rs7675290 is 5488 bp; rs8180214 is 4993 bp; rs8180209 is 4939 bp and rs17016071 is 4766 bp