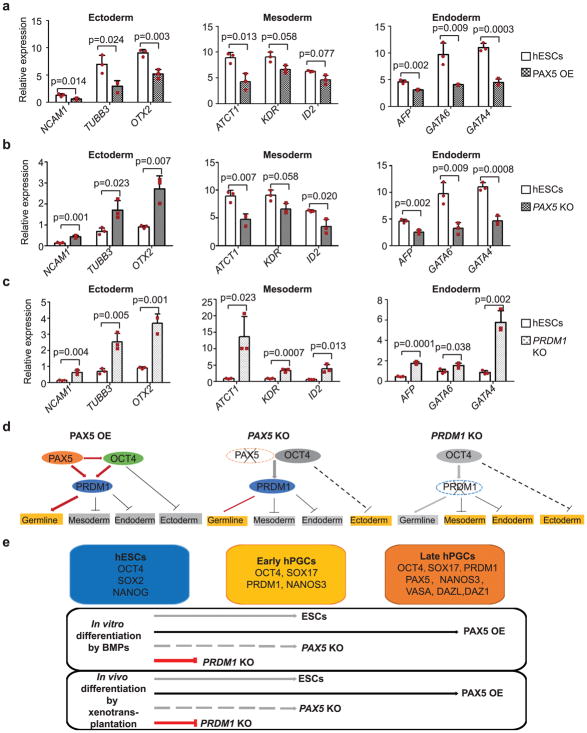
Figure 7. Role of PAX5 and PRDM1 in hPGC specification in vitro.
(a–c) RT-qPCR analysis of gene expression in all three germ layers in H1 hESCs, PAX5 OE cells (a) and H1 hESCs, PAX5 KO cells (b) and H1 hESCs, PRDM1 KO cells (c) after BMP induced differentiation. Data are represented as mean ± SD of n=3 independent replicates. P-values were calculated by two-tailed Student’s t-test. (d) Proposed molecular model for transcriptional network centered by PAX5, OCT4 and PRDM1 in hPGCs. Upon induced germ cell differentiation with BMPs, OCT4 expression is reduced to moderate levels and maintained in partnership with PAX5. To efficiently induce germline programs, OCT4 represses ectodermal genes and at the same time, together with PAX5, activates PRDM1 to repress mesodermal and endodermal genes. In PAX5 KO cells, OCT4 expression has decreased to levels so low that the expression of ectodermal genes has not been suppressed effectively. Thus, the efficiency of induction of germ cells is low in PAX5 KO cells and lower in PRDM1 KO cells: due to low expression of OCT4 and loss of PRDM1 function, genes in all somatic lineages are upregulated and germ cell programs fail to be activated. (e) Summary of data establishing roles of PAX5 and PRDM1 in hPGC specification in vitro and in vivo. The identity of hESCs is maintained by core transcriptional network centered by OCT4, SOX2 and NANOG. Induced by BMP signals in vitro or in vivo by xenotransplantation, hESCs start to differentiate to early hPGCs, which express early germ cell markers, such as OCT4, SOX17, PRDM1 and NANOS3 (Grey line with arrowhead); Overexpression of PAX5 is able to enhance the efficiency to early hPGCs and promote early hPGCs to the later stage, which express mature germ cell markers, such as DDX4, DAZL and DAZ1(Black line with arrowhead). Loss of PAX5 significantly reduces germ cell potential of hESCs (Grey dotted line with arrowhead), while loss of PRDM1 leads to failure of hPGC specification (red line with an end bar). Source data for a–c are in Supplementary Table 2.