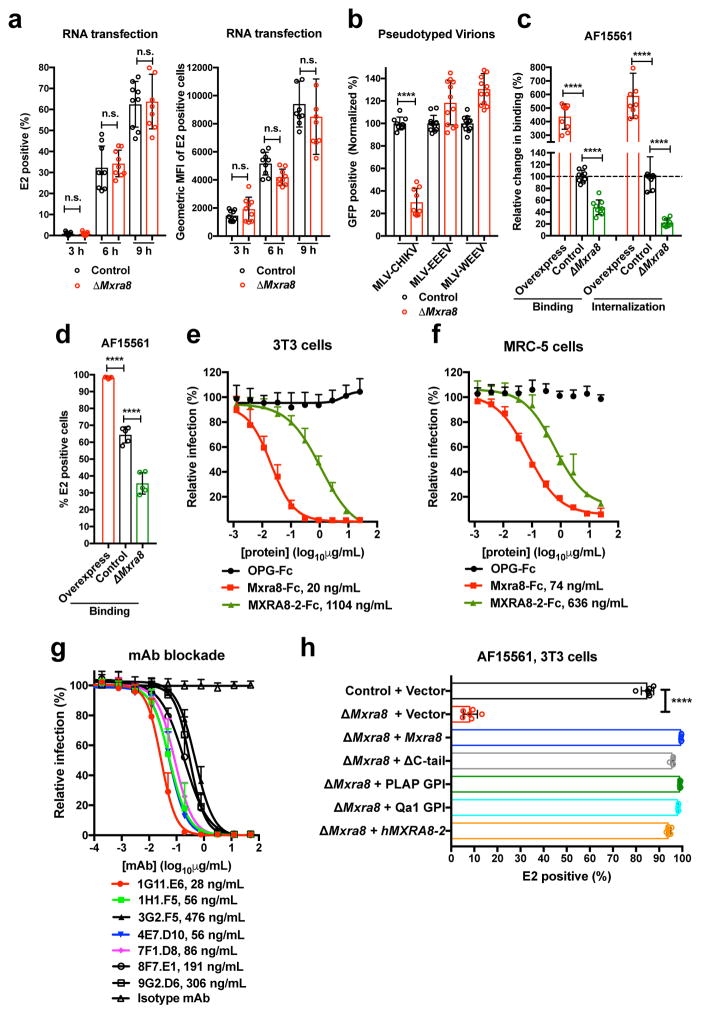
Figure 2. Mxra8 modulates CHIKV attachment and internalization.
a. Transfection of CHIKV RNA into control or ΔMxra8 cells. Cells were analyzed for E2 expression (left, percent positive; right, mean fluorescence intensity; 3 experiments, n = 9; not significantly different (n.s.), two-tailed t-test with Holm-Sidak correction, mean ± SD). b. MLV RNA encoding GFP and pseudotyped with alphavirus structural genes were added to control or ΔMxra8 cells. Data are from three (MLV-CHIKV, n = 9) or four (MLV-WEEV and MLV-EEEV, n = 12) experiments: (two-tailed t-test with Holm-Sidak correction, ****, P < 0.0001, mean ± SD). c–d. CHIKV-AF15561 was incubated with control, ΔMxra8, and Mxra8-overexpressing MEFs at 4°C (c, left and d) or 37°C (c, right) as described in Methods. Cells were harvested, and RNA (CHIKV and Gapdh) was measured by (c) RT-qPCR or surface E2 protein was analyzed by (d) flow cytometry (3 experiments, c, n = 9; d, n = 5; one-way ANOVA with Dunnett’s test, ****, P < 0.001; mean ± SD). e–f. Mxra8-Fc, MXRA8-2-Fc, or OPG-Fc were mixed with CHIKV-181/25 prior to infection of 3T3 (e) or MRC-5 (f) cells. Data are from two (MRC-5, n = 6) or three (3T3, n = 8–12) experiments: mean ± SD. g. Blockade of CHIKV-181/25 infection in 3T3 cells with hamster anti-Mxra8 or isotype control mAbs (2 experiments, n = 6; mean ± SD). h. Trans-complementation of ΔMxra8 cells with vector, Mxra8, Mxra8 ΔC-tail, Mxra8 with GPI anchors (PLAP or Qa1-derived), or human MXRA8-2 (3 experiments: (n = 6, one-way ANOVA with Dunnett’s test, ****, P < 0.001, mean ± SD).