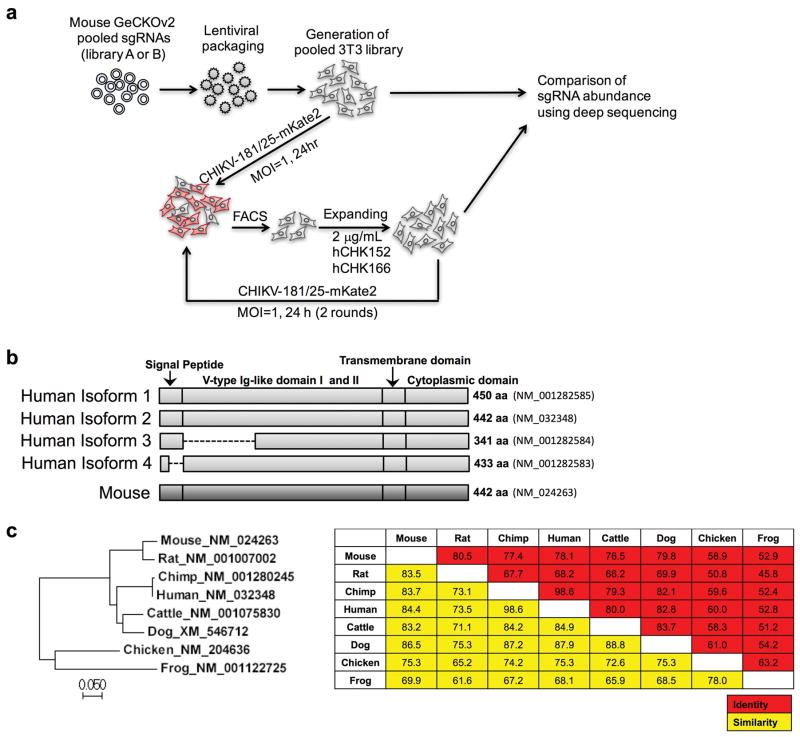
Extended Data Figure 1. RISPR-Cas9-based gene editing screen.
Mouse 3T3 cells were transduced separately with two half libraries (A + B) comprising 130,209 sgRNAs, selected with puromycin, and then inoculated with CHIKV-181/25-mKate2 (MOI of 1). One day later, mKate2-negative cells were sorted, and expanded in the presence of 2 μg/ml each of CHK-152 and CHK-166 neutralizing mAbs. Several days later, cells were re-inoculated with CHIKV-181/25-mKate2 without neutralizing mAbs and re-sorted for mKate2-negative cells. This procedure was repeated one additional time. Afterwards, genomic DNA was harvested for sgRNA sequencing and compared to the parent library for abundance (see Supplemental Tables 1 and 2). b. Diagram of the mouse Mxra8 and human MXRA8 orthologs. The transcript indentification numbers and length of proteins are indicated to the right. Partial deletions in the isoforms 3 and 4 are shown as dashed lines. c. Phylogenetic tree of Mxra8 indicating genetic relationships. The Neighbor-Joining tree was constructed using MEGA 7. Scale bar shows the branch length. (Right) Identity (red) and similarity (yellow) matrix indicating the conservation of Mxra8 between species. The matrix was generated using MagGat 1.8.