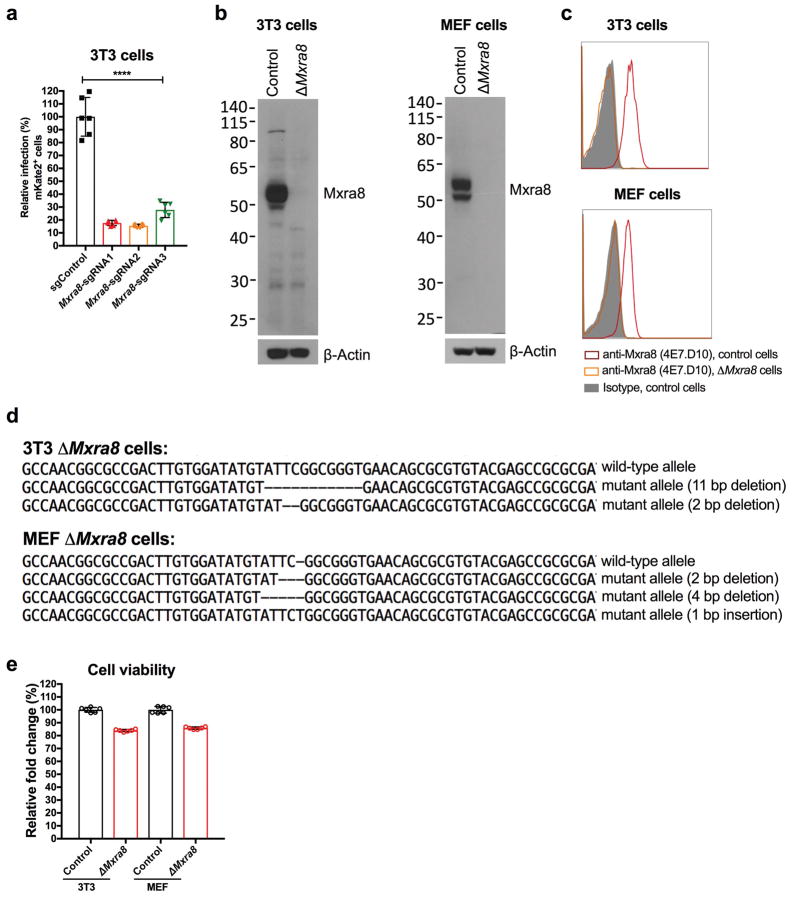
Extended Data Figure 2. Efficiency of targeting Mxra8 expression by CRISPR-Cas9 gene editing.
a. 3T3 cells were edited with a control or three different Mxra8 sgRNAs. After puromycin selection, bulk cells were inoculated with chimeric CHIKV-181/25-mKate2 and processed for marker gene expression by flow cytometry. Data are pooled from three experiments and expressed as mean ± SD (n = 6, one-way ANOVA with a Dunnett’s multiple comparison test compared to control, ****, P < 0.0001). b. Western blotting of Mxra8 in control and ΔMxra8 3T3 or MEF cells using hamster mAb 3G2.F5. One representative of two is shown. c. 3T3 and MEF cells (parent or ΔMxra8) were tested for Mxra8 surface expression by flow cytometry using anti-Mxra8 antibody (4E7.D10) and an isotype control mAb. One representative experiment of two is shown. d. Sanger sequencing of Mxra8 in control and ΔMxra8 3T3 or MEF cells. Sequencing data shows an alignment and individual out-of-frame deletions. e. Viability of control and ΔMxra8 3T3 and MEF cells. An equal number of cells were plated and viability was assessed over a 24 h period using the Cell-Titer Glo assay. The results were normalized to control cells and pooled from two experiments (n = 6). Error bars indicate SD.