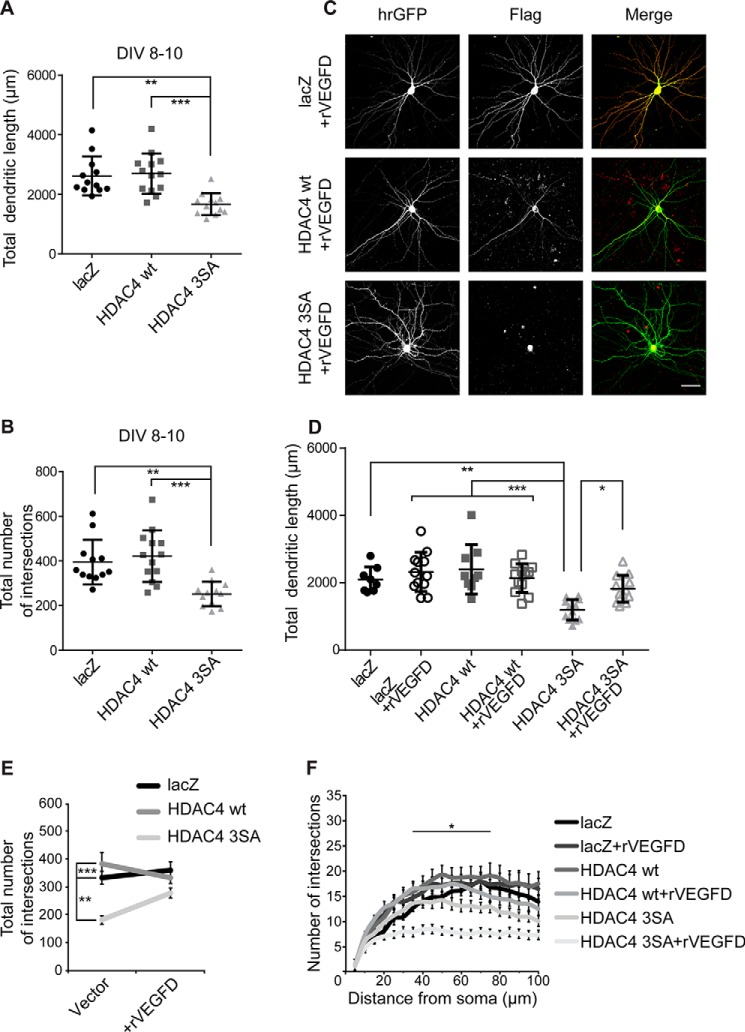
Figure 6.
VEGFD treatment prevents the simplification of dendritic morphology promoted by nuclear HDAC4. A, quantification of the total dendritic length of hippocampal neurons at DIV10. Cells were co-transfected with hrGFP and FLAG-tagged constructs lacZ, HDAC4 WT, and HDAC4 3SA at DIV8. HDAC4 3SA versus HDAC4 WT, p = 3 × 10−4; HDAC4 3SA versus lacZ, p = 1 × 10−3; lacZ versus HDAC4 WT, p = 0.94. B, total number of intersections derived from Sholl analysis at DIV10. HDAC4 3SA versus HDAC4 WT, p = 2 × 10−4; HDAC4 3SA versus lacZ, p = 2.1 × 10−3; lacZ versus HDAC4 WT, p = 0.76. C, representative images of cultured hippocampal neurons transfected as in A and treated or not with rVEGFD (100 ng/ml) for 3 days. FLAG tags were stained with Alexa 594. Scale bar is 40 μm. D, quantification of the total dendritic length of hippocampal neurons transfected as indicated, with or without treatment for 3 days with rVEGFD. HDAC4 3SA versus lacZ, p = 1.9 × 10−3; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA versus HDAC4 3SA + rVEGFD, p = 0.03; HDAC4 3SA + rVEGFD versus lacZ + rVEGFD, p = 0.14; HDAC4 3SA + rVEGFD versus HDAC4 WT + rVEGFD, p = 0.61. E, total number of intersections derived from the Sholl analysis shown in F. HDAC4 3SA versus lacZ, p = 2.2 × 10−3; HDAC4 3SA versus HDAC4 WT, p = 1 × 10−4; HDAC4 3SA + rVEGFD versus lacZ, p = 0.69; HDAC4 3SA + rVEGFD versus lacZ + rVEGFD, p = 0.15; HDAC4 3SA + rVEGFD versus HDAC4 WT, p = 0.06; HDAC4 3SA + rVEGFD versus HDAC4 WT + rVEGFD p = 0.57; lacZ versus lacZ + rVEGFD, p = 0.98; HDAC4 WT versus HDAC4 WT + rVEGFD, p = 0.76. F, Sholl analysis of hippocampal neurons transfected as indicated, with or without treatment for 3 days with rVEGFD. In total, 8 (lacZ), 9 (HDAC4 WT), and 12 neurons (lacZ + rVEGFD, HDAC4 WT + rVEGFD, HDAC4 3SA, HDAC4 3SA + rVEGFD) from 3 independent experiments were analyzed for each construct (D, E, and F). At DIV10, 12 (lacZ and HDAC4 3SA) and 13 (HDAC4 WT) neurons from 3 independent experiments were analyzed for each construct (A and B). Statistically significant differences were determined by one-way ANOVA (A, B, D, and E) and two-way ANOVA (F) followed by Tukey's post hoc test. ***, p < 0.001; **, p < 0.01; *, p < 0.05. For scatter plots, each point represents a value derived from one neuron. Graphs represent mean ± S.D.