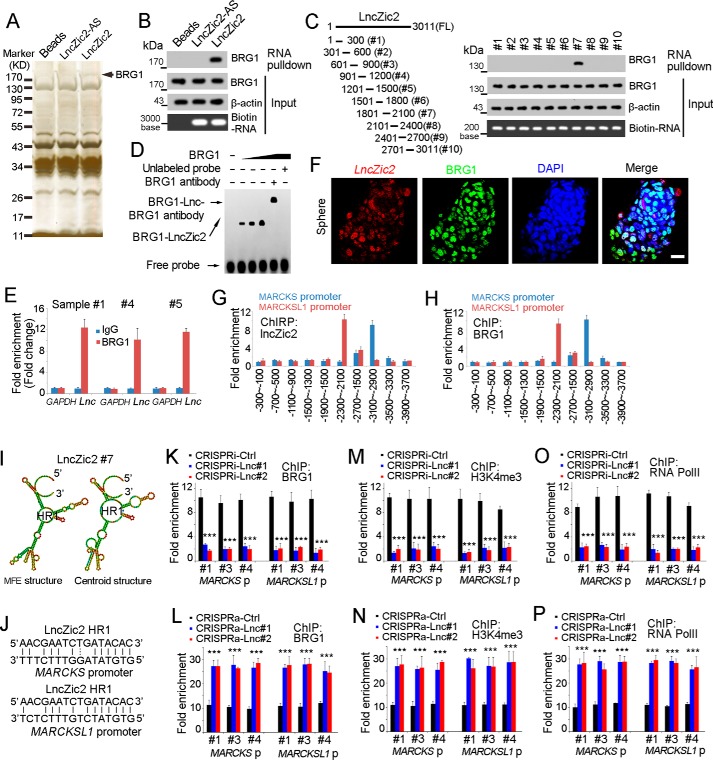
Figure 5.
lncZic2 recruited BRG1 to the MARCKS/MARCKSL1 promoter. A, biotin-labeled lncZic2 was generated through in vitro transcription and incubated with oncosphere lysate for RNA pulldown, and the specific band of lncZic2 was identified as BRG1 through mass spectrometry. B, RNA pulldown was performed, and the interaction between lncZic2 and BRG1 was confirmed by Western blotting. C, lncZic2 truncates were constructed (left panel), and an RNA pulldown assay was performed for BRG1 examination (right panel). FL, full length. HR, hairpin region. D, RNA EMSA confirmed the interaction between BRG1 and lncZic2. The #7 region of lncZic2 was used for RNA ESMA. E, RNA immunoprecipitation assays were performed with BRG1 antibody and control antibody, and the enrichment of lncZic2 was examined through real-time PCR. F, lncZic2 and BRG1 subcellular location was examined through FISH. Oncospheres were used for FISH, and co-localization of BRG1 and lncZic2 was observed. Scale bar = 10 μm. DAPI, 4′,6-diamidino-2-phenylindole. G and H, oncospheres derived from sample #1 were used for a ChIRP assay (G) or a ChIP assay (H), and the enrichment of the MARCKS/MARCKSL1 promoter was examined by real-time PCR. The indicated regions of the MARCKS/MARCKSL1 promoter were examined, and the data were normalized to lncZic2-AS probes (G) or IgG control antibody (H). I, the structure of the indicated region of lncZic2 was predicted through a tool available online (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party–hosted site.) MFE, minimum free energy. J, the sequence complementary between lncZic2 and MARCKS/MARCKSL1 promoter is shown. K and L, BRG1 ChIP assays were performed, and MARCKS/MARCKSL1 promoter enrichment was examined through real-time PCR. lncZic2-depleted cells (K) and lncZic2-overexpressing cells (L) were used for ChIP assays. Ctrl, control. M and N, lncZic2-depleted cells (M) and lncZic2-overexpressing cells (N) were used for H3K4Me3 ChIP, and the enrichment of the MARCKS/MARCKSL1 promoter was examined through real-time PCR. O and P, RNA polymerase II (RNA PolII) ChIP assays were performed using lncZic2-depleted cells (O) and lncZic2-overexpressing cells (P), followed by real-time PCR for enrichment of the MARCKS/MARCKSL1 promoter. Experiments were repeated at least three times. ***, p < 0.001 by t test.