ABSTRACT
Yellow fever (YF) is a reemerging public health threat, with frequent outbreaks prompting large vaccination campaigns in regions of endemicity in Africa and South America. Specific detection of vaccine-related adverse events is resource-intensive, time-consuming, and difficult to achieve during an outbreak. To address this, we have developed a highly transferable rapid yellow fever virus (YFV) vaccine-specific real-time reverse transcription-PCR (RT-PCR) assay that distinguishes vaccine from wild-type lineages. The assay utilizes a specific hydrolysis probe that includes locked nucleic acids to enhance specific discrimination of the YFV17D vaccine strain genome. Promisingly, sensitivity and specificity analyses reveal this assay to be highly specific to vaccine strain(s) when tested on clinical samples and YFV cell culture isolates of global origin. Taken together, our data suggest the utility of this assay for use in laboratories of varied capacity for the identification and differentiation of vaccine-related adverse events from wild-type infections of both African and South American origin.
KEYWORDS: yellow fever virus, 17D vaccine, diagnostic, real-time RT-PCR, locked nucleic acids
INTRODUCTION
Yellow fever virus (YFV) of the genus Flavivirus, family Flaviviridae, is mosquito-borne and endemic to tropical regions of Africa and South America. Clinical manifestations of yellow fever (YF) range from mild to severe and are marked by fever, jaundice, hemorrhage, shock, and death in 20 to 60% of severe cases (1). Historically, YF was a major public health threat from the 18th to the early 20th centuries in areas of North America, the Caribbean, and Europe, where YFV is no longer known to circulate. The geographic recession of YF from these areas is largely attributed to aggressive mosquito eradication and vaccination efforts (2, 3). In fact, the development of the YFV vaccine (YFV17D) (4, 5) was a landmark in vaccine history and, after decades of use, it is considered to be one of the most effective and safest vaccines available (6). Despite this, cases of multiorgan failure resembling wild-type YFV infection caused by overwhelming viremias following YFV vaccination have been documented (7, 8), while being exceedingly rare in their occurrence (9).
Because of the devastating and explosive nature of YF outbreaks, one confirmed disease case is identified as an emergency by the World Health Organization (10), and mass vaccination campaigns are considered in response in areas lacking sufficient vaccine coverage (1). As examples, vaccination campaigns targeting 30 million people in Angola, the Democratic Republic of the Congo, and Uganda were undertaken to stymie outbreaks of YF that occurred on the African continent in 2016 (11). During such campaigns, adverse outcomes following YFV vaccination are exceedingly difficult to accurately diagnose due to the circulation of wild-type YFV and the related occurrence of natural infections. In fact, current methods for YFV vaccine-related adverse event detection require advanced laboratory capabilities that are limited to regional reference laboratories. These methods include (i) YFV17D-specific monoclonal antibody detection by immunohistochemistry in postmortem tissue samples, (ii) isolation of YFV17D from clinical samples and detection with YFV17D-specific monoclonal antibodies, and/or (iii) amplification of YFV17D RNA from clinical samples and genome sequencing (12). Unfortunately, these techniques can take weeks to complete and need to be conducted by expert laboratorians. Because of this, real-time assays have been pursued as an attractive, rapid, and highly transferable alternative to these time-intensive methods for YFV17D-specific detection. To date, real-time reverse transcription-PCR (RT-PCR) assays have not been successful for the discrimination of YFV17D RNA from both South American and African lineage YFVs (13), largely due to the relatively conserved nature of the YFV17D genome compared to African lineage viruses (14).
To address limitations in real-time RT-PCR-based discrimination of YFV strains, locked nucleic acids (LNAs) are a promising tool for increasing primer sensitivity and specificity. The LNA base is incorporated into an oligonucleotide and induces a conformational change (15) that provides the LNA bases with enhanced binding strength for complementary bases (16) and greater mismatch discrimination (17). The incorporation of LNA bases into quantitative PCR (qPCR) probes has advanced the field of single nucleotide polymorphism (SNP) genotyping in allelic discrimination assays (18) and in hepatitis C virus genotyping (19). Promisingly, LNA-based-probe designs have been successful in the development of diagnostic tests that require discrimination between a single nucleotide mismatch, such as SNP genotyping (20), and diagnosis of bleeding disorders and sickle cell disease (21). Furthermore, LNAs have been used in the detection of pathogens and the characterization of antibiotic-resistant Mycobacterium tuberculosis in clinical samples (22). Moreover, the sensitivity and specificity of diagnostic real-time PCR assays for hepatitis B virus and Chlamydia suis have been improved by the use of LNA probes (23, 24).
In light of the above-described findings, we have investigated the utility of LNA bases, incorporated into YFV17D-specific real-time RT-PCR hydrolysis probes, for the specific differentiation of vaccine-related disease from naturally occurring YFV strains of global origin. As part of these investigations, eight SNPs were identified as being unique to the YFV17D genome compared to all other YFV lineages possessing a genetic description, and YFV17D-specific real-time RT-PCR probes were designed with LNA bases at these SNP locations accordingly. Candidate probes were then screened, and a preferred probe was selected due to its relative capacity for sensitivity and specificity. These efforts provided the foundation for the development of the presented YFV17D-specific real-time RT-PCR assay that incorporates a newly designed LNA probe to rapidly discriminate between YFV17D and wild-type South American and African YFV lineages.
MATERIALS AND METHODS
Unless otherwise noted, all procedures were performed in accordance with the manufacturers' protocols.
Viruses and RNA extraction.
All viruses were gown in Vero cells maintained with Dulbecco's modified Eagle's medium (DMEM; Gibco, Gaithersburg, MD) and supplemented with 2% fetal bovine serum (VWR, Radnor, PA, USA), sodium pyruvate (Gibco), sodium bicarbonate (Gibco), gentamicin (Biowhittaker, Walkersville, MD), and amphotericin B (HyClone, South Logan, UT). The viruses used in sensitivity and specificity analyses are listed in Table 1. One hundred forty microliters of virus supernatant was extracted using the QIAamp viral RNA kit (Qiagen, Valencia, CA) and eluted in 60 μl of AVE buffer (water, 0.04% sodium azide). All YFV work was performed according to biosafety level 3 safety requirements.
TABLE 1.
Yellow fever virus isolates used for specificity testing of the YFV17D-specific real-time RT-PCR assay
| Isolate name | Source | Yr | Location | Result (Cq)a |
|---|---|---|---|---|
| 17D-204 | Vaccine | Positive (22.6) | ||
| Asibi | Human | 1927 | Ghana | Negative |
| Couma | Human | 1961 | Ethiopia | Negative |
| DakAr 1279 | Mosquito | 1965 | Senegal | Negative |
| 14FA | Human | 1971 | Angola | Negative |
| 614819 | Human | 1974 | Panama | Negative |
| 1899/81 | Human | 1981 | Cusca District, Peru | Negative |
| BA-55 | Human | 1986 | Nigeria | Negative |
| BC-7914 | Human | 1993 | Baringo County, Kenya | Negative |
| BeH 622205 | Human | 2000 | Goias, Brazil | Negative |
| INS 382060 | Human | 2000 | Meta Department, Colombia | Negative |
| INHRR 7a-05 | Human | 2005 | Portuguesa, Venezuela | Negative |
| FVB 0196 | Human | 2006 | Cochabamba, Bolivia | Negative |
| FMD 1240 | Human | 2007 | Madre De Dios, Peru | Negative |
| CAREC M2-09 | Red howler monkey | 2009 | Trinidad | Negative |
| InHRR 10a-10 | Red howler monkey | 2010 | Monagas, Venezuela | Negative |
| MIS 1034b | Human | 2011 | Lima, Peru | Positive (18.9) |
Cq, quantification cycle.
Isolated from a vaccine-related adverse event.
Primers and probes.
All wild-type and vaccine 17D and 17DD YFV complete genomes available as of 9 May 2017 were downloaded from the National Center for Biotechnology Information nucleotide database (https://www.ncbi.nlm.nih.gov/nucleotide/) and were aligned using the Clustal W function of the MEGA 7 software (25). The alignments were evaluated for regions where 17D and 17DD genomes differed from all other lineages of YFV (see Fig. S1 in the supplemental material). Eight sets of primers and probes were designed from regions of SNPs between YFV vaccine genomes and all other YFV genomes. The probes (Integrated DNA Technologies, Coralville, IA) were designed to be specific to the vaccine genome with LNA bases incorporated at the SNP and adjacent nucleotides (Table S1). Additional LNA bases were added to the 3′-end of the probe to increase the melting temperature if necessary.
Standardization of YFV17D-specific real-time RT-PCR.
The YFV17D-specific real-time RT-PCR was standardized using the QuantiTect probe RT-PCR kit (Qiagen, Valencia, CA) with 1 μM each primer and 0.15 μM 6-carboxyfluorescein (FAM)-labeled probe with the following cycling conditions: 50°C for 30 min, 95°C for 15 min, 45 cycles of 94°C for 15 s, and 64°C for 1 min on the CFX96 real-time PCR detection system (Bio-Rad, Hercules, CA). The annealing temperature was optimized by running a gradient and testing YFV17D RNA and wild-type YFV Asibi RNA.
Ten microliters of extracted RNA was used in a final reaction volume of 25 μl. Each RNA sample was tested with a minimum of two technical replicates, and each assay included at least two no-template negative controls, negative extraction controls, YFV Asibi RNA-negative controls, and YFV17D RNA-positive controls.
Standard preparation for YFV17D-specific real-time RT-PCR assay.
YFV17D genomic RNA from cell culture supernatants was retrotranscribed using the SuperScript III first-strand synthesis kit (Life Technologies, Carlsbad, CA) and random hexamer primers. Target YFV17D cDNA was amplified using AccuPrime Taq DNA polymerase (Life Technologies) with 100 ng of cDNA and 10 μM primers (set 8, Table 2) under the following conditions: 94°C for 2 min; 35 cycles of 94°C for 30 s, 60°C for 30 s, and 68°C for 30 s; and a 4°C hold. The amplification product was checked for appropriate 200-bp size on a 1.2% E-gel (Life Technologies). Unpurified amplification products were cloned into the TOPO TA dual-promoter kit (Life Technologies). White-blue screening of transformed colonies was completed on imMedia Amp Blue plates (Life Technologies), and plasmid DNA was extracted using a miniprep kit (Qiagen, Valencia, CA). The YFV17D insert was verified using the sequencing primers provided in the TOPO TA kits, and BigDye Terminator sequencing (Life Technologies) was performed using the ABI 3130 instrument (Life Technologies).
TABLE 2.
Yellow fever virus 17D-specific primers and probe designed around single nucleotide variant in 17D genomes
| Primer (set 8) or probesa | Locationb | Sequence (5′ to 3′)c |
|---|---|---|
| Forward | 10312 | GTATTCTGTGGATGCTGACC |
| Reverse | 10412 | TATCCCGGTTTCAGGTTGTG |
| Probe (R) | 10331 | CTCACCCAGTTGCAG |
Probe has 5′,6-carboxyfluorescein and 3′-Iowa black FQ quencher. (R), reverse orientation.
Genome location based on 17D genome with GenBank accession no. NC_002031.
Bold type bases have locked nucleic acid chemistry. Underlined base represents a single nucleotide variant in the 17D genome compared to wild-type yellow fever viruses.
RNA copy controls were synthesized in vitro after linearization of the plasmid with 1 U KpnI-HF (New England BioLabs, Ipswich, MA) for 1 h at 37°C. Enzymatic reactions were cleaned up using the QIAquick PCR clean-up kit (Qiagen, Valencia, CA). RNA was synthesized using 500 ng of linearized plasmid in the mMessage mMachine T7 Ultra kit (Ambion, Austin, TX) and incubated at 37°C for 5 h, followed by Turbo DNase treatment. Transcribed RNAs were cleaned up using the RNeasy kit (Qiagen), and concentration was determined using NanoDrop One spectrophotometer (Thermo Fisher, Waltham, MA) and analyzed with the 2200 TapeStation Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA copy number was estimated using the following equation: number of copies = [RNA concentration (ng) × (6.022 × 1023)]/[length (bp) × (1 × 109) × 340] (SciencePrimer). Serial dilutions were made in molecular-grade water to generate a standard curve ranging from 1 × 105 copies/reaction to 1 copy/reaction.
Determination of analytical performance.
To determine the absolute limit of detection (LOD) of the YFV17D-specific assay, in vitro-transcribed RNA copy controls in the range of 1 ×105 copies/reaction to 1 copy/reaction were tested in two independent assays with six replicates each. The probability of detection for the 95% LOD was determined using a probit regression analysis in R. The standard deviations of the intra-assay quantification cycle (Cq) and coefficient of variation (CV) of the interassay variance were calculated in GraphPad Prism 7 (GraphPad Software, Inc., La Jolla, CA). The LOD of biological samples was determined by spiking normal human serum with YFV17D and serially titrating to generate a curve of 100 PFU/ml to 0.39 PFU/ml. RNA from dilutions was obtained by extracted 140 μl of spiked normal human serum using the QIAamp viral RNA kit (Qiagen, Valencia, CA) and eluting in 60 μl of AVE buffer. The 95% LOD was determined by probit regression in R, and the equivalent RNA copies were estimated by a standard curve.
The specificity of the assay was determined by testing for potential cross-reactivity with 10 μl of serially diluted RNA extracted from virus cell culture supernatants of different YFV isolates from all lineages (Table 1) and relevant cocirculating arboviruses in two independent assays. To ensure there were no PCR inhibitors in the extractions, RNA was serially diluted 10-fold from 1 × 100 to 1 × 10−6 in molecular-grade water prior to testing.
Validation of diagnostic YF17D real-time RT-PCR assay on clinically relevant samples.
All clinical samples tested in this study were fatal cases of YFV vaccine-related adverse events (n = 7) previously characterized by submission to the Arbovirus Disease Branch, Diagnostic and Reference Laboratory. Wild-type YFV-spiked serum samples were created by adding YFV cell culture fluid to normal human serum in concentrations ranging from 1 × 105 PFU/ml to 1 × 103 PFU/ml. All clinical and spiked samples were extracted using the QIAamp viral RNA kit (Qiagen, Valencia, CA) by extracting 140 μl of serum and eluting in 60 μl AVE buffer. All samples were tested in duplicate in the standardized protocol described above using 10 μl of RNA. Samples were also tested in parallel with pan-yellow fever-specific primers (YFall) (26), with an annealing temperature of 60°C.
RESULTS
Standardization and selection of YFV17D-specific real-time RT-PCR assay.
All eight sets of primers and probes designed around the eight single nucleotide changes in YFV17D genome were tested against YFV17D RNA extracted from cell culture supernatant, with an annealing temperature (Ta) gradient ranging from 55°C to 70°C. Primer set 8 (Table 2) displayed positive amplification from 55°C to 69°C and produced the most efficient reaction, with ending relative fluorescence unit (RFU) values higher than the other primer sets at all Ta tested. Primer set 8 also had the lowest Cq (18.84) at the standard Ta of 60°C and was therefore chosen for further evaluation.
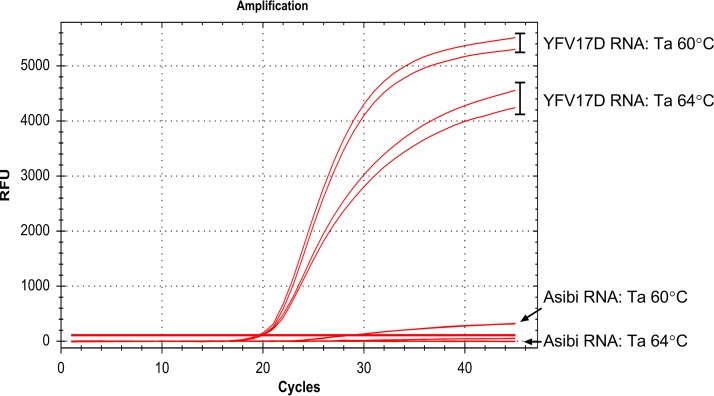
Initial cross-reactivity was tested using YFV Asibi RNA with YFV17D RNA as a positive control at a Ta of 60°C, which produced the best amplification curves in the gradient test. Primer set 8 had positive amplification with YFV17D (Cq, 19.03) with a high ending RFU value (5,400 RFU) (Fig. 1). Primer set 8 also had positive amplification with Asibi RNA (Cq, 26.14), with a substantially lower ending RFU (385 RFU). Finally, we investigated the higher Ta range of primer set 8 in determining specificity by testing YFV17D and YFV Asibi RNA at a Ta of 64°C (Fig. 1). Primer set 8 had positive amplification of YFV17D RNA (Cq, 19.29). Primer set 8 did not amplify YFV Asibi RNA at a Ta of 64°C, while primer set 8 at a Ta of 60°C did display positive amplification. Based upon these tests, primer set 8 was chosen for all further evaluation based on the reaction efficiency measured by ending RFU values when tested against YFV17D RNA, and the Ta was set to 64°C based on the lack of amplification when tested against YFV Asibi RNA. These standardized procedures were applied to all additional evaluations and referred to as the YFV17D-specific assay.
FIG 1.
Standardization and specificity of yellow fever 17D vaccine virus-specific locked nucleic acid hydrolysis probes. Real-time RT-PCR locked nucleic acid probes were designed to be specific to a single nucleotide polymorphism identified in the yellow fever virus (YFV) vaccine genome. The specificity of primer set 8 was optimized by testing RNA from YFV strains 17D (vaccine) and Asibi (parental strain) at an annealing temperature (Ta) of 60°C or 64°C. RFU, relative fluorescence unit.
Evaluation of the YFV17D-specific real-time RT-PCR assay.
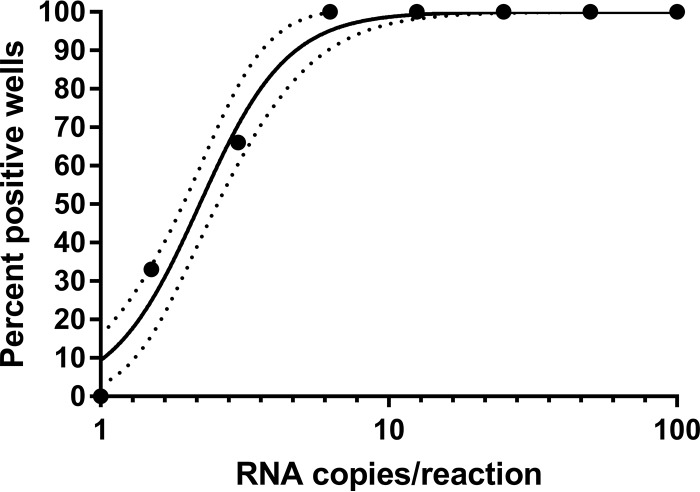
The analytical LOD and intra- and interassay precision were assessed by evaluating serial dilutions of RNA genome copy controls. Probit regression analysis determined the 95% LOD to be 6.2 RNA copies per reaction (95% confidence interval [CI], 3.9 to 8.7 copies per reaction) (Fig. 2). The calculated intra-assay precision of the YFV17D-specific real-time RT-PCR for replicates of serial diluted RNA copies ranged from 0.18 to 2.5 Cq standard deviations, with the greatest dispersion for the lower RNA copy load samples near the LOD. The calculated interassay precision ranged from CV of 0.4% to 3.5%, with the greatest CV for the lowest RNA copy load samples near the LOD. The LOD was also determined on biologically relevant samples by using serial titrations of YF17D spiked into normal human serum. The 95% LOD of the extracted RNA was found to be 1.7 PFU/ml, which corresponded to 10.12 RNA copies when plotted on a standard curve.
FIG 2.
Limit of detection analysis of yellow fever 17D vaccine virus-specific real-time RT-PCR assay. Probit regression analysis of the yellow fever 17D-specific assay was performed at an annealing temperature of 64°C using serial dilutions of in vitro transcribed RNA copies. Solid circles represent positive test results. Dotted lines represent the 95% confidence interval. Cq, quantification cycle.
The specificity of the YFV17D-specific assay was determined by testing 10 μl of serially diluted RNA from cell culture supernatant of wild-type YFV of all lineages and cocirculating arboviruses. The YFV17D-specific assay was highly specific and produced negative results for all wild-type YFV isolates tested (Table 1), with the exception of MIS 1034, which was identified as an isolate from a vaccine-related adverse event and was detected, with a Cq value of 18.94 on undiluted RNA. Negative test results were also obtained when the YFV17D-specific assay was tested with dengue viruses 1 to 4, West Nile virus, chikungunya virus, St. Louis encephalitis virus, and Zika virus.
Application of the YFV17D-specific real-time RT-PCR assay to clinically relevant samples.
The high specificity of the assay on RNA from cell culture supernatants led us to use the assay on clinical samples from vaccine-related adverse events. The YFV17D-specific assay was tested in parallel with pan-YFV primers (the YFall primers, previously described in reference 26), on samples from fatal vaccine-related adverse events (n = 7). Both assays were able to detect YFV17D RNA in all vaccine-related adverse event samples tested (Table 3). These data suggest that the YFV17D-specific assay is comparably sensitive in clinical settings.
TABLE 3.
Specificity of yellow fever virus 17D vaccine real-time RT-PCR assay on clinically relevant samples
| Virusc | Sample type | PFU/ml | Result (Cq)a |
|
|---|---|---|---|---|
| YFV17D | YFallb | |||
| 17D vaccine | Serum | Positive (34.6) | Positive (35.0) | |
| Serum | Positive (35.8) | Positive (36.2) | ||
| Serum | Positive (32.2) | Positive (33.6) | ||
| Lung | Positive (25.8) | Positive (28.3) | ||
| Liver | Positive (30.1) | Positive (31.3) | ||
| Kidney | Positive (26.4) | Positive (27.7) | ||
| Thymus | Positive (29.3) | Positive (29.9) | ||
| BA-55 | Spiked serum | 100,000 | Negative | Positive (26.0) |
| 10,000 | Negative | Positive (27.2) | ||
| 1,000 | Negative | Positive (30.4) | ||
| FMD 1240 | Spiked serum | 10,000 | Negative | Positive (22.9) |
| 1,000 | Negative | Positive (26.4) | ||
| INHRR 10a-10 | Spiked serum | 100,000 | Negative | Positive (20.9) |
| 10,000 | Negative | Positive (23.3) | ||
| 1,000 | Negative | Positive (25.3) | ||
Normal human serum spiked with wild-type YFV (n = 8) were tested in parallel with the YFV17D-specific assay and the YFall assay (26). The YFV17D-specific real-time RT-PCR assay did not amplify any wild-type YFV, while the YFall primers detected RNA in all samples tested (Table 3). These data suggest that the YFV17D-specific assay is specific to YFV17D in clinically appropriate sample types. Taking into account the low Cq values obtained in the YFall assay, the YFV17D-specific assay was unable to detect wild-type virus in the presence of high viral and RNA loads, further suggesting specificity.
DISCUSSION
Here, we describe the first report of a real-time RT-PCR assay for the specific discrimination of YFV17D among both South American and African lineage wild-type YFV strains. According to our analyses, the specificity of the assay is promising for the global detection of YFV vaccine-related adverse events and is attributed to the use of LNA bases in the oligonucleotide probe at an YFV vaccine SNP location. While our study benefited from testing clinical samples of YFV vaccine-related adverse events, unfortunately, the validation of the presented assay was limited by the lack of availability of wild-type YFV clinical samples. In light of this shortcoming, we utilized low-passage-number YFV isolates obtained from human infections and tested RNAs from both high-titer tissue culture fluid, as well as spiked human serum to verify specificity. In light of our findings, while continued validation is warranted, the YFV17D-specific assay can be used as a rapid clinical diagnostic tool in YFV outbreaks globally to assist epidemiological investigations and decision-making during mass vaccination campaigns in South America and Africa.
When designing the presented assay, two major considerations were taken into account. First, the increased binding strength of LNAs, which causes a high affinity for LNA-LNA duplexes and therefore the possibility of primer-dimer formation (27–29), was a major consideration. For this reason, we chose a low probe concentration of 0.15 μM to limit the formation of primer-dimers. Accordingly, the effect of probe concentration on potential dimer formation needs to be taken into consideration when this assay is validated in individual laboratories. The standard use of negative extraction and no-template controls is recommended at all times to facilitate the identification of erroneous results potentially attributed to dimer formation and contamination, respectively. Second, we compared all published YFV wild-type, 17D-204, and 17DD genome sequences with isolation dates from 1927 to 2017 to ensure that targeted SNP sites are consistent across all 17D and 17DD sequenced vaccine stocks (30, 31) and distinguish the vaccine stocks from all wild-type isolates across geographic locations. Our in silico analyses (Fig. S1) suggest that the YFV17D-specific assay is specific to 17D and 17DD genomes, while all past and current wild-type YFV sequences, including primary clinical sequences, maintain the SNP site. While virus isolation and genome sequencing are still vitally important for monitoring the genetic stability of the vaccine and confirming diagnosis, the YFV17D-specific assay presented here will enable more labs with a rapid diagnosis of YFV vaccine-related adverse events and provide information for selective virus isolation and sequencing of positive samples at regional reference labs.
Finally, we recommend that the presented YFV17D-specific assay be utilized in parallel with, or secondary to, a global YFV real-time RT-PCR diagnostic assay, such as those previously described by others (26, 32). Screening all samples with a global YFV real-time RT-PCR assay would ensure that only YFV-positive samples are tested to reduce unnecessary testing and improve resource conservation, and to allow for labs to have flexibility in implementing the YFV17D assay in routine YFV diagnostics and surveillance. By using this two-step diagnostic approach, positive amplification with a global YFV assay would verify infection, and, when accompanied with positive amplification in the YFV17D-specific assay, would verify the presence of YFV17D vaccine RNA in the determination of an adverse event.
Supplementary Material
ACKNOWLEDGMENTS
We thank Brad Biggerstaff of the CDC for assistance in statistical analysis, Robert Lanciotti of the CDC for technical review, and Erin Staples of the CDC and Robert Downing of Uganda Virus Research Institute (UVRI) for assistance in sample contribution.
The mention of trade names or products is solely for the purpose of providing specific information and does not imply endorsement by the CDC. The findings and conclusions in this paper are those of the authors and do not necessarily represent the views of the CDC.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.00323-18.
REFERENCES
- 1.Monath TP, Vasconcelos PF. 2015. Yellow fever. J Clin Virol 64:160–173. doi: 10.1016/j.jcv.2014.08.030. [DOI] [PubMed] [Google Scholar]
- 2.Soper FL. 1963. The elimination of urban yellow fever in the Americas through the eradication of Aedes aegypti. Am J Public Health Nations Health 53:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frierson JG. 2010. The yellow fever vaccine: a history. Yale J Biol Med 83:77–85. [PMC free article] [PubMed] [Google Scholar]
- 4.Theiler M, Smith HH. 1937. The use of yellow fever virus modified by in vitro cultivation for human immunization. J Exp Med 65:787–800. doi: 10.1084/jem.65.6.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Theiler M, Smith HH. 1937. The effect of prolonged cultivation in vitro upon the pathogenicity of yellow fever virus. J Exp Med 65:767–786. doi: 10.1084/jem.65.6.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Menezes Martins R, Fernandes Leal Mda L, Homma A. 2015. Serious adverse events associated with yellow fever vaccine. Hum Vaccin Immunother 11:2183–2187. doi: 10.1080/21645515.2015.1022700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martin M, Tsai TF, Cropp B, Chang GJ, Holmes DA, Tseng J, Shieh W, Zaki SR, Al-Sanouri I, Cutrona AF, Ray G, Weld LH, Cetron MS. 2001. Fever and multisystem organ failure associated with 17D-204 yellow fever vaccination: a report of four cases. Lancet 358:98–104. doi: 10.1016/S0140-6736(01)05327-2. [DOI] [PubMed] [Google Scholar]
- 8.Vasconcelos PF, Luna EJ, Galler R, Silva LJ, Coimbra TL, Barros VL, Monath TP, Rodigues SG, Laval C, Costa ZG, Vilela MF, Santos CL, Papaiordanou PM, Alves VA, Andrade LD, Sato HK, Rosa ES, Froguas GB, Lacava E, Almeida LM, Cruz AC, Rocco IM, Santos RT, Oliva OF, Brazilian Yellow Fever Vaccine Evaluation Group. 2001. Serious adverse events associated with yellow fever 17DD vaccine in Brazil: a report of two cases. Lancet 358:91–97. doi: 10.1016/S0140-6736(01)05326-0. [DOI] [PubMed] [Google Scholar]
- 9.Lindsey NP, Rabe IB, Miller ER, Fischer M, Staples JE. 2016. Adverse event reports following yellow fever vaccination, 2007–13. J Travel Med 23:. doi: 10.1093/jtm/taw045. [DOI] [PubMed] [Google Scholar]
- 10.WHO. 1998. District guidelines for yellow fever surveillance. Division of Emerging and other Communicable Diseases, Surveillance and Control, World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/64384/www9834.pdf?sequence=1. [Google Scholar]
- 11.World Health Organization. 2016. Yellow fever situation report: 28 October 2016. World Health Organization, Geneva, Switzerland: http://www.who.int/emergencies/yellow-fever/situation-reports/28-october-2016/en/. [Google Scholar]
- 12.World Health Organization. 2008. Detection and investigation of serious adverse events following yellow fever vaccination: guidance from an informal consultation of experts, 18–19 November 2008, Geneva, Switzerland. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/handle/10665/70251/WHO_HSE_GAR_ERI_2010.2_eng.pdf?sequence=1. [Google Scholar]
- 13.Fischer C, Torres MC, Patel P, Moreira-Soto A, Gould EA, Charrel RN, de Lamballerie X, Nogueira RMR, Sequeira PC, Rodrigues CDS, Kummerer BM, Drosten C, Landt O, Bispo de Filippis AM, Drexler JF. 2017. Lineage-specific real-time RT-PCR for yellow fever virus outbreak surveillance, Brazil. Emerg Infect Dis 23:. doi: 10.3201/eid2311.171131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hahn CS, Dalrymple JM, Strauss JH, Rice CM. 1987. Comparison of the virulent Asibi strain of yellow fever virus with the 17D vaccine strain derived from it. Proc Natl Acad Sci U S A 84:2019–2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaur H, Arora A, Wengel J, Maiti S. 2006. Thermodynamic, counterion, and hydration effects for the incorporation of locked nucleic acid nucleotides into DNA duplexes. Biochemistry 45:7347–7355. doi: 10.1021/bi060307w. [DOI] [PubMed] [Google Scholar]
- 16.Wengel J, Petersen M, Nielsen KE, Jensen GA, Hakansson AE, Kumar R, Sorensen MD, Rajwanshi VK, Bryld T, Jacobsen JP. 2001. LNA (locked nucleic acid) and the diastereoisomeric alpha-l-LNA: conformational tuning and high-affinity recognition of DNA/RNA targets. Nucleosides Nucleotides Nucleic Acids 20:389–396. doi: 10.1081/NCN-100002312. [DOI] [PubMed] [Google Scholar]
- 17.You Y, Moreira BG, Behlke MA, Owczarzy R. 2006. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res 34:e60. doi: 10.1093/nar/gkl175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson MP, Haupt LM, Griffiths LR. 2004. Locked nucleic acid (LNA) single nucleotide polymorphism (SNP) genotype analysis and validation using real-time PCR. Nucleic Acids Res 32:e55. doi: 10.1093/nar/gnh046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davalieva K, Kiprijanovska S, Plaseska-Karanfilska D. 2014. Fast, reliable and low cost user-developed protocol for detection, quantification and genotyping of hepatitis C virus. J Virol Methods 196:104–112. doi: 10.1016/j.jviromet.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Mouritzen P, Nielsen AT, Pfundheller HM, Choleva Y, Kongsbak L, Moller S. 2003. Single nucleotide polymorphism genotyping using locked nucleic acid (LNA). Expert Rev Mol Diagn 3:27–38. doi: 10.1586/14737159.3.1.27. [DOI] [PubMed] [Google Scholar]
- 21.Ugozzoli LA, Latorra D, Puckett R, Arar K, Hamby K. 2004. Real-time genotyping with oligonucleotide probes containing locked nucleic acids. Anal Biochem 324:143–152. doi: 10.1016/j.ab.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Zhao Y, Li G, Sun C, Li C, Wang X, Liu H, Zhang P, Zhao X, Wang X, Jiang Y, Yang R, Wan K, Zhou L. 2015. Locked nucleic acid probe-based real-time PCR assay for the rapid detection of rifampin-resistant Mycobacterium tuberculosis. PLoS One 10:e0143444. doi: 10.1371/journal.pone.0143444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lis P, Kumala A, Spalinski M, Rypula K. 2014. Novel locked nucleic acid (LNA)-based probe for the rapid identification of Chlamydia suis using real-time PCR. BMC Vet Res 10:225. doi: 10.1186/s12917-014-0225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q, Wang X, Zhang J, Song G. 2012. LNA real-time PCR probe quantification of hepatitis B virus DNA. Exp Ther Med 3:503–508. doi: 10.3892/etm.2011.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Domingo C, Patel P, Yillah J, Weidmann M, Mendez JA, Nakoune ER, Niedrig M. 2012. Advanced yellow fever virus genome detection in point-of-care facilities and reference laboratories. J Clin Microbiol 50:4054–4060. doi: 10.1128/JCM.01799-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ballantyne KN, van Oorschot RA, Mitchell RJ. 2008. Locked nucleic acids in PCR primers increase sensitivity and performance. Genomics 91:301–305. doi: 10.1016/j.ygeno.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 28.McTigue PM, Peterson RJ, Kahn JD. 2004. Sequence-dependent thermodynamic parameters for locked nucleic acid (LNA)-DNA duplex formation. Biochemistry 43:5388–5405. doi: 10.1021/bi035976d. [DOI] [PubMed] [Google Scholar]
- 29.Owczarzy R, You Y, Groth CL, Tataurov AV. 2011. Stability and mismatch discrimination of locked nucleic acid-DNA duplexes. Biochemistry 50:9352–9367. doi: 10.1021/bi200904e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stock NK, Boschetti N, Herzog C, Appelhans MS, Niedrig M. 2012. The phylogeny of yellow fever virus 17D vaccines. Vaccine 30:989–994. doi: 10.1016/j.vaccine.2011.12.057. [DOI] [PubMed] [Google Scholar]
- 31.Beck A, Tesh RB, Wood TG, Widen SG, Ryman KD, Barrett AD. 2014. Comparison of the live attenuated yellow fever vaccine 17D-204 strain to its virulent parental strain Asibi by deep sequencing. J Infect Dis 209:334–344. doi: 10.1093/infdis/jit546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weidmann M, Faye O, Faye O, Kranaster R, Marx A, Nunes MR, Vasconcelos PF, Hufert FT, Sall AA. 2010. Improved LNA probe-based assay for the detection of African and South American yellow fever virus strains. J Clin Virol 48:187–192. doi: 10.1016/j.jcv.2010.04.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.