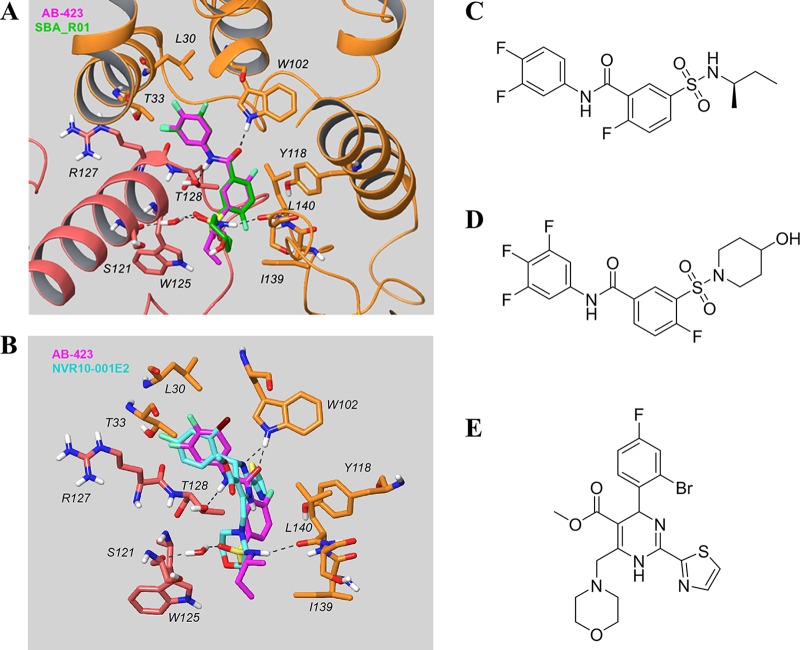
FIG 9.
In silico docking of AB-423 into the X-ray structure of core protein. (A) Comparison of the predicted binding model of AB-423 and the X-ray structure of SBA_R01 (PDB accession number 5T2P; resolution, 1.69 Å) bound to the dimer-dimer interface of the Cp-Y132A viral core protein. Key residues in the vicinity of the ligand binding site are shown (the carbon of the capsid protein is in orange for chain B and pink for chain C, the carbon of the ligands is in magenta for AB-423 and green for SBA_R01, with nitrogen in blue, sulfur in yellow, oxygen in red, and fluorine in light green). The other residues within contact distance were not displayed for clarity. (B) Comparison of the predicted binding model of AB-423 and the X-ray structure of NVR10-001E2 (PDB accession number 5E0I; resolution, 1.95 Å) bound to the dimer-dimer interface of the Cp-Y132A viral core protein. Key residues in the vicinity of the ligand binding site are shown (the carbon of the capsid protein is in orange for chain B and pink for chain C, the carbon of the ligands is in magenta for AB-423 and cyan for NVR10-001E2, with nitrogen in blue, sulfur in yellow, oxygen in red, fluorine in light green, and bromine in dark brown). The other residues within contact distance are not displayed for clarity. (C to E) Chemical structures of AB-423 {(R)-5-[N-(sec-butyl)sulfamoyl]-N-(3,4-difluorophenyl)-2-fluorobenzamide} (C), SBA_R01 {4-fluoro-3-[(4-hydroxypiperidin-1-yl)sulfonyl]-N-(3,4,5-trifluorophenyl)benzamide} (D), and NVR10-001E2 [methyl 6-(2-bromo-4-fluorophenyl)-4-(morpholinomethyl)-2-(thiazol-2-yl)-1,6-dihydropyrimidine-5-carboxylate] (E).