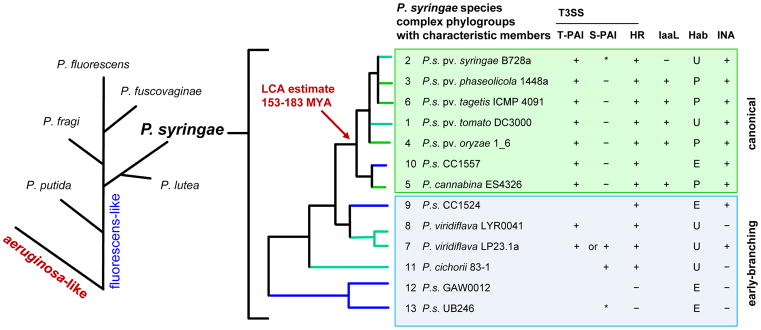
Figure 1. The phylogeny of P. syringae and common phylogroup features.
On the left, proposed phylogenetic branching order for major species groups within the P. fluorescens-like major branch of Pseudomonas14, 15. On the right, thirteen identified phylogroups (PGs) in the P. syringae species complex, based on multi-locus sequence analysis (MLSA). Phylogroups representing monophyletic species within the complex are noted. Characteristic PG members are listed along with general phylogroup-associated features when known16. S-PAI, single-part pathogenicity islands lack a canonical CEL but may carry CEL T3SS effectors within the hrp/hrc cluster. IaaL, presence of the indole acetic acid lysine synthetase gene for the inactivation of auxin29. Hab, common habitat, strains are isolated mostly from plants (P) or the environment (E), or both/ubiquitous (U). INA, reported ice nucleation capacity or the presence of the inaW ice-nucleation gene. IaaL, Hab and INA traits vary on a strain-to-strain basis. *, PG 2 clade c and PG 13 have A-typical S-PAIs (A-PAI) with distinct genomic locations12.