Figure 1. FIGURE 1: Evolution of the SVCT clade in NATs.
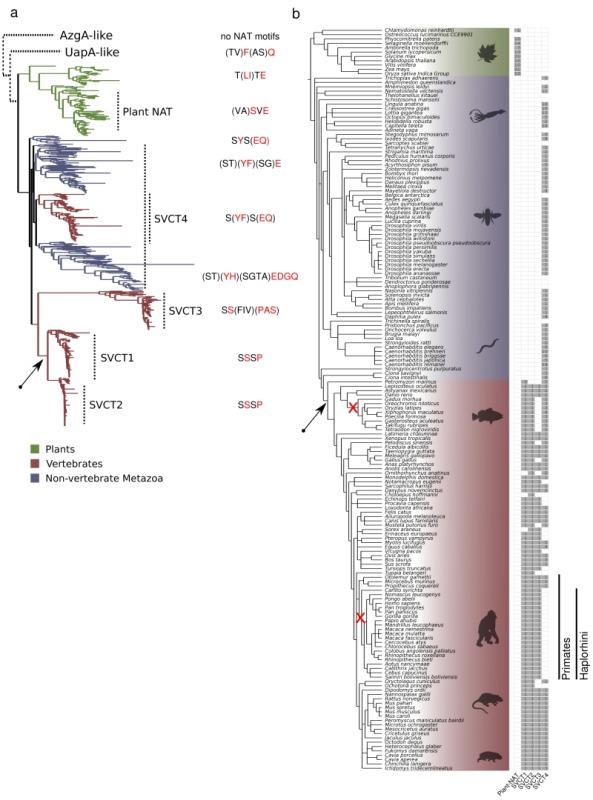
(a) ML phylogenetic tree of the SVCT clade of NATs and the distinct NAT signature motifs. The different SVCT plant and animal NAT subfamilies, as well as a variable consensus of the four first amino acids in the NAT signature motif, are indicated next to the tree. The tree was midpoint rooted, as shown, and the other two clades, UapA-like and AzgA-like, are placed schematically above with dotted lines. The UapA-like group corresponds to canonical microbial NATs and is shown in more detail in Supplementary Figure S1. The AzgA-group is used as an out-group of structurally homologous proteins, which however do not conserve the functional motifs of canonical NATs. For the complete version of the plant and animal NATs see also Supplementary Figure S2.
(b) Phylogenetic profile of the SVCT clade sub-families for the 171 plant and animal species. The frequency of members in each subfamily is shown next to the NCBI taxonomy tree structure of the 171 species that were searched for NAT domains. The sub-families were defined based on (a) and in reference to the known mammalian transporters (SVCT1-4). Two early losses in two lineages, the SVCT3 in the Teleostean clade Eurypterygii and SVCT4 in the primate suborder Haplorhini, are indicated in the corresponding branches with a red "x". The emergence of the L-ascorbic acid transport specificity is indicated with a black arrow both in the phylogenetic tree (a), as well as in the taxonomy tree (b). The colors (green, red, blue) in both (a) and (b) are as indicated in the figure legend. Both trees were visualized using the ETE toolkit 42.
