Figure 6.
Full-Length mRNAs Can Be Upregulated upon Exosome Depletion
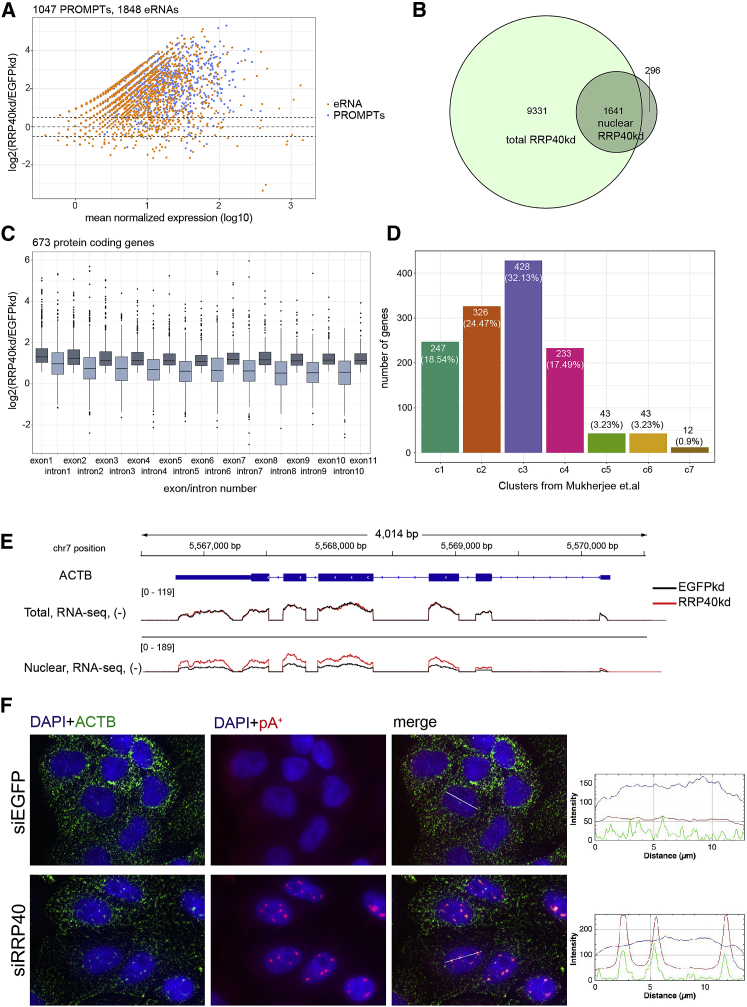
(A) Log2 fold changes (y axis) between PROMPTs and eRNAs in nuclear RRP40kd versus EGFPkd libraries plotted against their mean normalized expression (log10 scale, x axis).
(B) Venn diagram representing the overlap between significantly upregulated protein-coding genes (log2[RRP40kd/EGFPkd] > 0, padj < 0.05) in the nuclear RNA libraries and the upregulated and unaffected protein-coding genes (log2[RRP40kd/EGFPkd] > −0.5) in the total RNA libraries.
(C) Boxplots of log2 fold changes (RRP40kd/EGFPkd) of each individual exon and intron of the selected protein coding genes in ascending order up to the 11th exon.
(D) Bar plots representing the classification of 1,641 protein-coding genes—overlap from (B)—to previously described clusters (Mukherjee et al., 2017). Note the enrichment of genes belonging to cluster c3 (Pearson’s chi-square test, p = 6.12e−10; calculated against all expressed genes in nuclear samples represented in Figure S6E).
(E) Genome browser view of the ACTB locus, showing tracks of normalized total and nuclear RNA-seq data from control (EGFPkd, black line) and RRP40-depleted (red line) HeLa cells. All tracks represent an average of three biological replicates, except the nuclear EGFPkd sample, which represents average of two biological replicates.
(F) Dual ACTB and pA+ RNA-FISH analysis in the indicated factor-depleted HeLa cells. Merged images of DAPI (blue), ACTB (green), and pA+ RNA (red) are shown. Image display is as in Figure 1C.
See also Figure S6E.