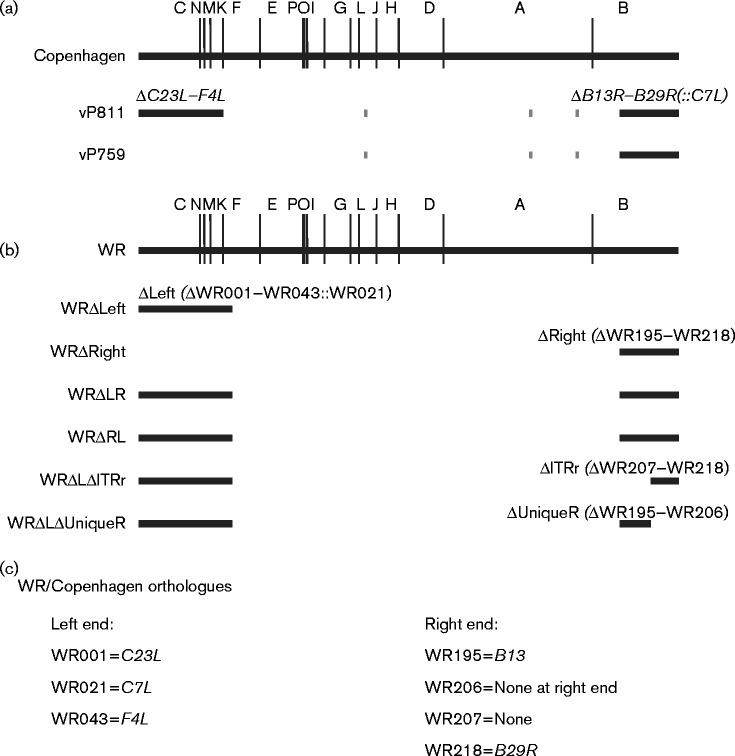
Fig. 1.
Position of large deletions in recombinant viruses. (a) HindIII map of the VACV Copenhagen genome, showing the position of the deletions at the termini of the genome in either vP811 or vP759. Short grey lines represent the thymidine kinase gene, A-type inclusion locus region and haemagglutinin gene that are deleted from both strains. The gpt selectable marker is present at the left end of vP811 and the right end of vP759. The human host-range gene C7L is present at the right terminus of vP811. (b) HindIII map of the VACV WR genome, showing the position of the deletions at the termini of the genome in six of the recombinant viruses produced. The EGFP/bsd fusion gene and VACWR021 (C7L) are present at the left end of all recombinant viruses carrying ΔLeft, and the mCherry gene was inserted at the right end to select for deletions ΔRight, ΔITRr or ΔUniqueR. (c) WR and Copenhagen orthologues relevant to (a, b). The genes delimiting the large deletions are the same across VACV strains, but some of the genes within these blocks differ due to strain differences.