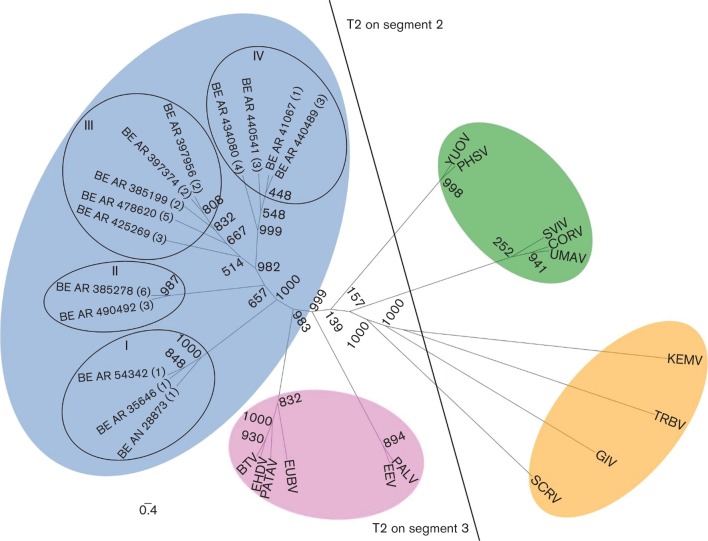
Fig. 4.
Relationships between complete amino acid sequences of T2 proteins of different orbiviruses. Maximum-likelihood unrooted tree showing distinct phylogenetic groups based on the T2 protein. Group ‘T2 on segment 3′ includes viruses in which the T2 protein [in this case VP3 (T2)] is encoded on segment 3: Culicoides-borne (pink) and sandfly-borne (blue) viruses composed of different serotypes of the Changuinola group. Group ‘T2 on segment 2’ includes viruses in which the T2 protein [in this case VP2 (T2) or VP3 (T2)] is encoded on segment 2: mosquito-borne (green) and tick-borne (orange). Bootstrap values are placed at each main node of the tree. Roman numerals (I to IV) represent the phylogenetic lineages. Arabic numerals (1 to 5) correspond to different geographical locations from where the viruses were isolated. (1) Belém–Brasilia highway km 94, Pará state; (2) Monte Dourado/Jari, Pará state; (3) base 4/Tucuruí, Pará state; (4) Serra Norte/Carajás, Pará state; (5) Balbina/Pte. Figueiredo, Amazonas state; (6) Porto Trombetas, Pará state. Full names of virus isolates and accession numbers of T2 protein sequences used for comparative analysis are listed in Table S4. The scale bar represents 0.4 % genetic divergence.