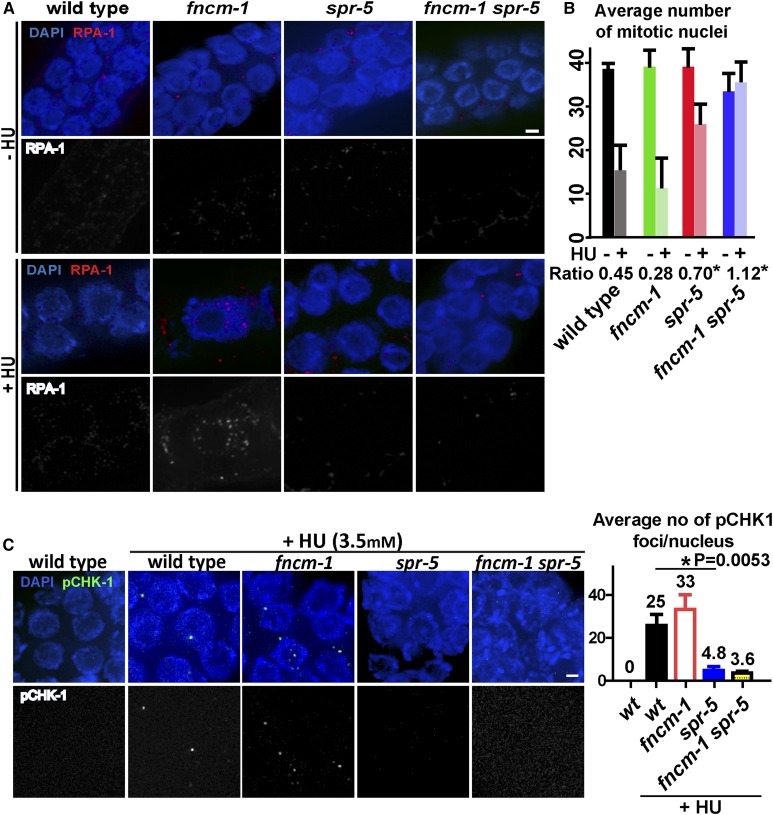
Figure 3.
FNCM-1 promotes replication-fork progression and SPR-5 is required for the S-phase checkpoint sensing the single-stranded DNA region formed upon lack of FNCM-1. (A) Immunolocalization of single-stranded DNA binding protein RPA-1 upon 3.5 mM HU treatment in the PMT for the indicated genotypes. Bar, 2 µm. (B) Quantitation of the average number of mitotic nuclei within 40 µm in the PMT region of the germlines from the indicated genotypes. Ratio represents the number of nuclei observed following HU treatment (+HU) divided by the number observed without treatment (−HU). * indicates statistical significance compared to wild-type control. P = 0.0422 for spr-5, P = 0.0095 for fncm-1 spr-5. P-values calculated by the two-tailed Mann–Whitney U-test, 95% C.I. (C) S-phase DNA damage checkpoint activation is impaired in spr-5 single and fncm-1 spr-5 double mutants. Left: Immunostaining for pCHK-1 on germline nuclei at the PMT following 3.5 mM HU treatment. Bar, 2 µm. Right: Quantitation of pCHK-1 foci. n = 4–6 gonads. P-values calculated by the two-tailed Mann–Whitney U-test, 95% C.I.