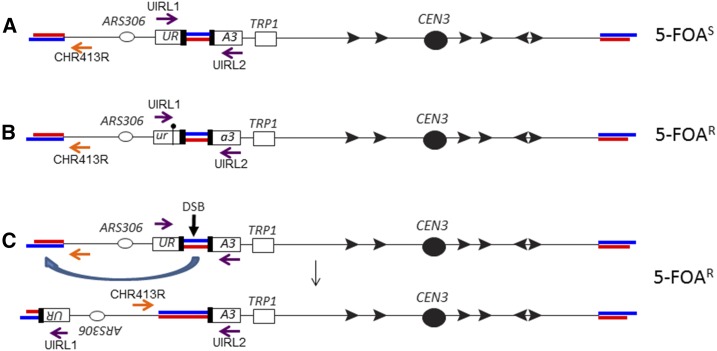
Figure 1.
Genetic system used to detect genomic alterations induced by ITSs. Telomeres and ITSs are shown as paired red and blue lines, with red and blue representing the CA-rich and GT-rich strands, respectively. The top strand is shown with the 5′ end on the left side. Only telomeric repeats with the GT-rich strand on the 3′ end have telomere function. Black arrows indicate Ty elements, and purple and orange arrows indicate the orientations and positions of primers used to diagnose genomic alterations. The ITS is replicated by forks initiated at ARS306. The figure is not drawn to scale. (A) Reporter gene used in our study (Aksenova et al. 2013). The ITSs are inserted within an intron embedded with URA3; the length of the ITS is 120 bp, representing 15 copies of an 8-bp repeat (TGTGTGGG). Strains with this reporter are Ura+ Trp+, and we selected 5-FOAR derivatives to detect genomic alterations. (B) Point mutations induced by the ITSs. These derivatives are Ura− Trp+ and have a PCR fragment of the expected size when the purple primers flanking the ITS are used (UIRL1 and UIRL2 in Table S2). Sequencing of PCR fragments demonstrates that these events are associated with a point mutation in the URA3 coding sequences flanking the ITS. (C) Terminal inversions induced by the ITSs. In this class, no PCR fragment is found with the purple primers, but a PCR fragment is observed using the orange primer (Chr3_413R in Table S2) and the centromere-proximal purple primer (UIRL2). In addition to the 5-FOAR phenotype, most of this class are Trp−, as a consequence of epigenetic silencing of URA3 caused by the longer region of telomeric repeats resulting from the inversion; the Trp− phenotype is reversed when the cells are analyzed on plates lacking tryptophan but containing the Sir2p-inhibitor nicotinamide. 5-FOAR, 5-FOA-resistant; DSB, double-strand break; ITS, interstitial telomeric sequence.