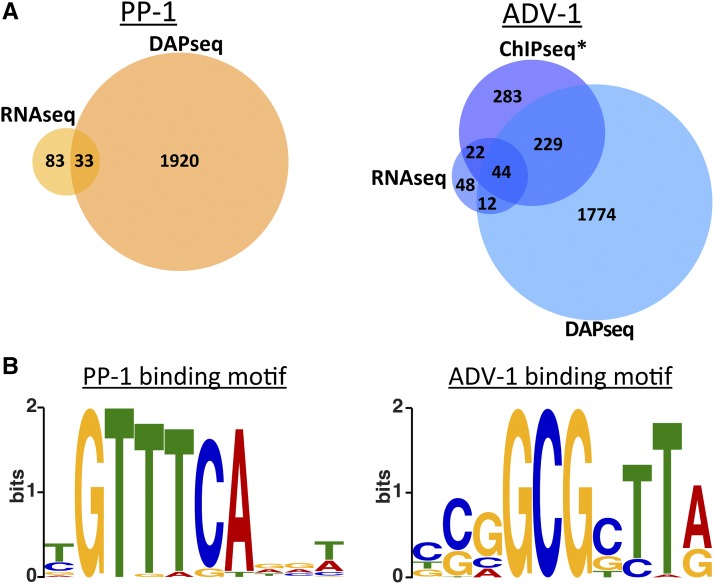
Figure 3.
DAP-seq identifies promoters bound by ADV-1 or PP-1. (A) Number of genes that are downregulated in Δpp-1 germlings (RNA-seq) or bound by PP-1 (DAP-seq) (left panel). The number of genes that are down regulated in Δadv-1 germlings (RNA-seq) or bound by ADV-1 (DAP-seq and ChIP-seq) (right panel). Downregulated genes were identified by consensus between Cuffdiff, EdgeR, and DESeq2, log2 fold change < −2 and adjusted P-value <0.01 (compare with Figure 1C). Genes bound by each transcription factor were counted if the transcription factor was bound within 2 kb upstream of the ATG (P < 0.001). *ADV-1 ChIP-seq data are available from Table S3 in Dekhang et al. (2017), in which ChIP-seq was performed at four different circadian time points (24 hr dark, 24 hr dark + 15 min light, 24 hr dark + 30 min light, and 24 hr dark + 60 min light). Genes were included here if they were bound by ADV-1 during at least one time point. (B) Consensus DNA binding motif for PP-1 or ADV-1 based on DAP-seq data.